STOECKERT S.#, PODKOLODNAYA O.A., KEL A.E., BRUNK B.*, HAAS J.*, SALAS F.*, STEPANENKO I.L., IGNATIEVA E.V., KEL-MARGOULIS O.V., ANANKO E.A., PODKOLODNY N.L., OVERTON G.C.*, KOLCHANOV N.A.
Institute of Cytology and Genetics, (Siberian Branch of the Russian Academy of Sciences), 10 Lavrentieva ave., Novosibirsk, 630090 Russia e-mail: opodkol@bionet.nsc.ru
#Division of Hematology, The Children’s Hospital of Philadelphia, Abramson Center, 34th and Civic Center Bllvd., Philadelphia, PA 19104-4318, USA
*Department of Genetics, U. of Pennsylvania School of Medicine, Room 475, Clinical Research bilding, 422 Curie Boulevard , Philadelphia, PA 19104-6145, USA
Keywords: database, gene expression, ontogenesis, differentiation, erythroid cell, protein, mRNA, developmental stages, expression dynamics, transcription factors, growth factors
Summary
The EpoGERD database collects information on features of gene expression in erythropoietic tissues, cells, or cell lines. Structure of the database and the means for its integration with other databases are described. The developed format providing the experimental data formalized representation is detailed
Introduction
The data on structure, function, and expression of eukaryotic genes are being accumulated rapidly. A great number of molecular biological databases have been developed aiming to systematize and accumulate these data in a user-friendly format. The largest databases of general orientation – EMBL[Rodriguez-Tome P. et al., 1996], GenBank [Benson D.A. et al., 1996], PIR [George D.G. et al., 1996], and SWISS-PROT [Rodriguez-Tome P. et al., 1996] – compile the information on DNA, RNA, and protein sequences as well as certain structure-function characteristics of genes and proteins. More specialized databases deal with either particular aspects or particular objects of molecular biological investigations [Kel, A.E.,et al 1997][Wingender et al., 1996] [Ringwald et al., 1997] [Westerfield et al., 1997], [Baxevanis et al., 1998]. [Salas et al., 1998]. The data on gene structures that are necessary for specialized databases can be partially extracted from the general databases, such as EMBL and GenBank, whereas only the original publications can be the source of information on gene expression patterns and regulation.
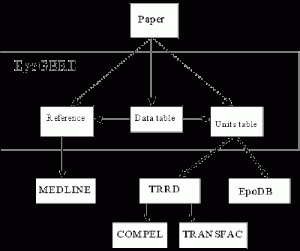
Fig. 1. Distribution of input information flow from an individual publication in the EpoGERD database.
The goal of this work was to develop a computer database for compiling the data on expression patterns of vertebrate genes and regulation of their expression in the course of ontogenesis and differentiation of the erythroid cell in a format convenient for automatic processing. The format of the database EpoGERD (Gene Expression Regulation Database) and functional characterization of the main gene groups presently available in the database are described.
Format of the database
Only the experimentally substantiated information on gene expression in erythropoietic tissues, erythroid cells, and cell lines and regulation of their expression extracted from original publications is included into the EpoGERD database. In the current version of EpoGERD, all information is accumulated in three tables: Reference table, Expression Units table, and Data table. The distribution of input information flow from an individual publication in the EpoGERD database is shown in Fig. 1. Bibliographic information is stored in the Reference table, which includes index number ascribed to each paper, complete citation information, and its MEDLINE accession number.
Table 1. Example of the entry in the EpoGERD Expression Units table
| Record | Content |
| T00311 | Index number of expression unit in the EpoGERD database |
| erythropoietin | Full name of expression unit |
| Epo | Short name of expression unit |
| mouse, Mus musculus | Species |
| 293402, 36289 | Accession number of the corresponding gene in EpoDB |
| G001221 | Accession number of the corresponding gene in TRRD |
The information on the protein or mRNA isoform described in a publication is filled in the Expression Units table. One gene can produce several isoforms of mRNA ( protein) that are frequently expressed in a different tissue- and development-specific manner. The Expression Units table of EpoGERD implies that the names of all protein and mRNA isoforms originating form one gene are included; that is, the expression patterns of each mRNA isoform or the corresponding protein can be described in the EpoGERD. For each entry, the Expression Units table indicate the short and full names of expression unit and its synonyms, species of the organism expressing the unit described, its EpoGERD index number, and accession numbers of the gene coding for this expression unit in the EpoDB [Salas 1998] and TRRD [Kel, A.E. et al., 1997] databases. An example of the entry in the EpoGERD Expression Units table is shown in the table 1.
Thus, one record in the Expression Units table contains the information for identification of both the gene and expression unit as well as allows the cross-references to EpoDB and TRRD. The Expression Units Table provides also for the connection with the databases COMPEL [Kel, O.V. et al 1997] and TRANSFAC [Heinemeyer et al 1998] due to the unified gene numbering in TRRD, COMPEL, and TRANSFAC (Fig. 1).
The results of individual experiments on gene expression and its regulation described in original publications are stored in the Data table. Each entry in this table corresponds to a separate experiment described in a separate publication; it describes an individual expression pattern of an individual transcription unit and may contain a considerable amount of data. The format of the database allows accumulation of both qualitative and quantitative data.
The links of the Data table with Expression Units and Reference tables is performed due to the index number of expression units and original papers indicated in each entry of this table (Fig. 1). The data obtained in the experiments on both the mRNA and protein levels is included into the database; the method used to obtain these data on expression regulation is indicated obligatory. All this information is pooled in a joint informational module. This module is connected with the other module compiling the information on conditions of the experiments and the experimental data themselves. The format of the database considers the following conditions of the experiments:
- developmental stages, at which gene expression was studied;
- cell cycle stages, at which gene expression was studied;
- the types of tissue, in which gene expression was studied;
- cells and cell types, in which gene expression was studied;
- external factors, the influence of which were studied (effects of several factors simultaneously can be considered as well as the time of their action).
Both individual and tabulated data can be inputted; the latter are useful to follow the time dynamics of gene expression. In case the input data are tabulated, the expression dynamics is shown as a graphical profile.
Table 2. Example of an individual entry of the Data table
| Field | Contents |
| EX_AC | E00643 |
| EX_DT | 29/10/97 |
| EX_TYP | RNA |
| EX_NAME | slot blot analysis |
| DA_AC | E00643.D06 |
| DA_TR | T00204; mouse, Mus musculus; c-fos |
| DA_DEV | postnatal:8 weeks |
| DA_IND | erythropoietin |
| DA_CON | 200 mU/mL |
| DA_IND | chemical:AZT |
| DA_CON | 10 uM |
| DA_CEL | committed progenitor cells:erythroid |
| DA_TIS | bone marrow |
| DA_STR | mouse strain: CD-1 |
| DA_INI | start of culture |
| DA_QNT | hours |
| DA_QNT_UNIT | % of control |
| DA_QNT_VAR | None |
| QNT_TAB | 24, 97 |
| REF_ID | L000173 |
The record in the field EX_AC (Table 2) indicates the index number of this experiment in the Data table: E00643; in the EX_DT, the date when this experiment was added to the table. The fields EX_TYP and EX_NAME explain that the level of mRNA was studied in this experiment by slot blot analysis; DA_TR, that it was a mouse c-fos mRNA that has an ascribed number T00204 in the Expression Units table. The field DA_DEV indicates that the experimental samples were taken from animals eight weeks postnatum; fields DA_IND and DA_CON, that the studies were carried out on the background of erythropoietin and AZT at concentrations of 200 mU/ml and 10 m M, respectively. Records in the fields DA_CEL, DA_TIS, and DA_STR describe that the experiment was performed employing erythroid committed progenitor cells obtained from bone marrow of CD-1 mice. The time in the experiment was recorded in hours from the start of culture, as indicated in the fields DA_INI and DA_QNT. The records in the fields DA_QNT_UNIT and QNT_TAB explain that the expression is recorded as % of the control and amounts to 97% 24 h after the effect of two external factors.
The format described provides the possibility to describe qualitative, semiquantitative, and quantitative data on expression patterns in different organs, tissues, cells, and cell lines as well as to compile the information on gene expression dynamics in the course of ontogenesis, cell differentiation, and at different stages of the cell cycle. In addition, the format of the database allows the consideration of the effect of external factors and signals on gene expression level.
A number of vocabularies are supported within the EpoGERD database to unify the records inputted to the Data table, in particular, vocabularies of experimental methods, organs, tissues, cells and cell lines, animal lines and cell strains, developmental stages, measurement units, transforming agents, and graphical profiles of expression dynamics. The vocabularies form the basis for further automatic analysis of the data accumulated.
Contents of the EpoGERD database
The EpoGERD database contains currently the data on 168 expression units expressed at various stages of vertebrate ontogenesis in erythropoietic tissues, cells, or cell lines. The species represented are listed in Table 3.
The genes described in the EpoGERD database are expressed mainly in hematopoietic cells and their precursors. Among them are mRNAs and proteins expressed in a wide range of tissues including derivatives of housekeeping genes (88) and mRNA with tissue-specific expression manner (80).
The EpoGERD database was designed to compile the information on expression patterns of the genes during ontogenesis and cell differentiation. Selection of the genes to be described in the EpoGERD database aims to represent both the genes that are regulated during the cell maturation and the genes encoding the products that determine the direction of this regulation. The major functional groups of genes in the EpoGERD database are listed in Table 4
Table 3. The species represented in the EpoGERD database
| Species |
Number of expression units |
| Mouse |
54 |
| Human |
69 |
| Chicken |
24 |
| Rat |
2 |
| Frog |
13 |
| Hamster |
6 |
Table 4. Functional groups of genes represented in the EpoGERD database
| Expression units |
Number |
| Structural and transport proteins |
33 |
| Enzymes |
17 |
| Transcription factors |
49 |
| Cell cycle regulatory proteins |
15 |
| Growth factors and their receptors |
16 |
| Others |
38 |
Conclusion
Thus, the database described here allows the accumulation of a diverse information concerning patterns and regulation of gene expression. Analyses of the data compiled in the EpoGERD database will aid the estimation of time dynamics and the order of expression switch-on of the genes encoding the products that determine morphological and functional specificity of the cell; investigation of the genes coordinately expressed during ontogenesis and differentiation of cells and tissues; and study the effects of external stimuli on sets of coordinately expressed genes.
A diverse and multilevel interactions between the products of activated genes can be represented as a gene network, a challenging problem of modern molecular biology and bioinformatics. Accumulation and cataloging of large massifs of data on gene expression patterns and functional interactions is one of the necessary conditions for the progress in this direction [Bryant et al., 1998 ]. The database format described is sufficiently unified and allows the accumulation of the data on genes involved in other functional systems.
Acknowledgments
This work was partially supported by the Russian State Human Genome Program (12312 GCh-5) and Russian Foundation for Basic Research (grants 98-04-49479, 97-04-49740 and 96-04-50006), U.S. National Institute of Health (2-R01-RR02026-08A2)
References
- Baxevanis, A.D., Landsman, D., Histone Sequence Database: new histonefold family members, Nucleic Acids Res., 261, 372-375 (1998).
- Benson D.A., Boguski M., Lipman D.J., Ostell J. GenBank. Nucl. Acids Res. 24, 1-5 (1996)
- Beroud C., Virder F., Soussi T. p53 gene mutation: software and database. Nucl. Acids Res. 24, 147-150 (1996)
- Bryant, B., Milosavljevic, A., Somogyi, R., Gene expression and genetic networks, Proceedings of the Pacific Symposium on Biocomputing, P. 3 (1998).
- Calligaris, R., Bottardyi, S., Cogoi, S., Apezteguia, I., Santoro, C., Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor, Proc. Natl. Acad. Sci. USA, , 92., P. 11598-11602. (1995)
- Chretien, S., Dubart, A., Beaupain, D., Raich, N., Grandchamp, B., Rosa, J., Goossens, M., Romeo, P.H., Alternative transcription and splicing of the human porphobilinogen deaminase gene result either in tissue-specific or in housekeeping expression, Proc. Natl. Acad. Sci. USA, 85, P. 6-10 (1988).
- George D.G., Barker W.C., Mewes H.W., Pfeffer F., Tsugita A. The PIR-International protein sequence database Nucl. Acids Res. 24, 17-20 (1996)
- Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A. E., Kel, O.V., Ignatieva, E.V., Ananko, E.A., Podkolodnaya, O.A., Kolpakov, F.A., Podkolodny N.L., Kolchanov, N.A., Databases on Transcriptional Regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res., 26, 364-370. (1998)
- Kel, A.E., Kolchanov, N.A., Kel, O.V., Romashchenko ,A.G., Anan`ko, E, Ignat`eva, E.V., Merkulova, T.I., Podkolodnaya, O.A., Stepanenko, I.L., Kochetov, A.V, Kolpakov, F.A., Podkolodny, N.L., Naumochkin, A.N., TRRD: Database on Transcription Regulatory Regions of Eukaryotic Genes., Mol. Biol. (Mosk,), 31, No. 4, p. 626-636, (1997).
- Kel, O.V., Kel, A.E., Romashchenko, A.G., Wingender, E., Kolchanov, N.A., Composite regulatory elements: Classification and description in the COMPEL database, Mol. Biol. (Mosk,), 31, No. 4, p. 601 (1997).
- Michel, G.S., Carr, D.B., Askenazi, M., Fuhrman, S., Wen, X., Cluster analysis and data visualization of large-scale gene expression data. Proceedings of the Pacific Symposium on Biocomputing, P. 42. (1998).
- Pischedda, C., Cocco, S., Melis, A., Marini, M.G., Kan, Y.W., Cao, A., Moi, P., Isolation of a differentially regulated splicing isoform of human NF-E2, Proc. Natl. Acad. USA, 92, 3511, (1995).
- Ringwald, M., Davis, G.L., Smith, A.G., Trepanier, L.E., Begley, D.A., Richardson, J.E., Eppig, J.T., The mouse gene expression database GXD, Seminars in Cell and Developmental Biology, 5, 489-497 (1997).
- Rodriguez-Tome P. , Stoehr P.J., Cameron G.N., Floers T.P. The european bioinformatics institute (EBI) databases. Nucl. Acids Res. 24, 6-12 (1996)
- Salas, F., Haas, J., Brunk, B., Stoecker, Jr., C.J., Overton, G.C., EpoDB: a database of genes expressed during vertebrate erythropoiesis. Nucleic Acids Res., 26, P. 288-289 (1998).
- Westerfield, M., Doerry, E., Kirkpatrick, A.E., Driever, W., Douglas, S.A., An on-line database for zebrafish development and genetics research, Seminars in Cell and Developmental Biology, 5, 477-488 (1997).
- Wingender, E., Dietze, P. , Karas, H., Knuppel, R., TRANSFAC: a database on transcription factors and their DNA binding sites, Nucl. Acids Res., 1996, 24, P. 238-241.