KOLCHANOV N.A., IGNATIEVA E.V., KEL-MARGOULIS O.V., KEL A.E., ANANKO E.A., PODKOLODNAYA O.A., STEPANENKO I.L., MERKULOVA T.I., GORYACHKOVSKY T.N., KOLPAKOV F.A., PODKOLODNY N.L., LAVRYUSHEV S.V., GRIGOROVICH D.A., FROLOV A.S., ROMASHCHENKO A.G.
Institute of Cytology and Genetics, (Siberian Branch of the Russian Academy of Sciences), 10 Lavrentieva ave., Novosibirsk, 630090 Russia
+Corresponding author e-mail:kol@bionet.nsc.ru
Keywords: transcription, regilatory regions, database, regulation, eukaryotic genes, sites, transcription factors, expression pattern, promoters, enhancers, silencers
Current state of the TRRD database is described including the new possibilities of experimental data formalized representation connected with TRRD format modification. Structure of the database and the means for its integration with other databases are detailed. Contents of the TRRD, Release 4.0, are briefly analyzed.
Introduction
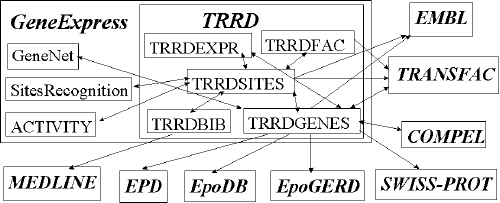
The Transcription Regulatory Regions Database (TRRD) is a convenient tool for studying the complex process of transcription regulation. The model of structure-function organization of transcription regulatory regions of eukaryotic genes [A.E. Kel et al., 1997; O.V. Kel et al., 1995] that formed the basis of the TRRD database takes into consideration a great diversity of elements involved in transcription control, their block structure, and the hierarchy essential for their functioning. At present, the TRRD database contains five interconnected tables (Fig. 1): TRRDGENES (general description of genes), TRRDEXP (description of expression patterns), TRRDSITES (description of sites), TRRDFAC (description of transcription factors), and TRRDBIB (references to original papers). The information on expression patterns of individual genes, structure of their regulatory regions, and transcription factors involved in transcription control comes from reviewing original publications. The TRRD is installed under the SRS (http://sgi.sscc.ru/), providing easy information retrieval and integration with other databases and computer systems for data processing. The TRRD database is a part of the global system GeneExpress [Kolchanov et al., 1998a] available at Molecular Biological Server of the Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences (http://wwwmgs.bionet.nsc.ru/).
Fig. 1 Structure of the TRRD database and its links to other databases and computer systems.
Format of the TRRD database
A modified version of the format that allows a more precise presentation of information in a computer-readable form was used in the latest TRRD Release 4.0. The format of TRRD (releases 2.1 and 3.3) were described earlier [A.E. Kel’ et al., 1997; O.V. Kel et al., 1995]. Thus, here we present the formats of the tables TRRDEXP and TRRDFAC, absent in the earlier releases and supplement to the table TRRDGENES on hypersensitivity sites.
Description of expression patterns (Table TRRDEXP). A number of fields characterizing the gene expression pattern are used to describe the conditions under which a gene is expressed. The expression pattern implies a set of data on dependence of gene expression on (1) cell cycle stage; (2) developmental stage; (3) cell type, tissue, or organ; and (4) influence of signals external to the cell: chemicals, cytokines, hormones, growth factors, etc., and the level of gene expression under these conditions in qualitative and quantitative terms. Fig. 2a illustrates the gene expression pattern description on the example of rat acyl-coenzyme A synthetase gene. One of the major advantages of the new scheme implemented in Release 4.0 is in the presence of internal links between structural regulatory units and a set of data on gene expression pattern. For example (Fig. 2a), the fenofibrate-induced increase in the rat acyl-coenzyme A synthetase gene (G001195) expression in adult animal liver involves the binding site AN 2397 located in the promoter AN P00543 (a fragment of the binding site AN 2397 description is shown in Fig. 2b).
|
The description of the gene expression pattern |
|
| RE G001195.019 | “Expression vector” identifier |
| RT mRNA | mRNA content was studied in the experiment |
| RD adult | Developmental stage of the organism under experiment |
| RO liver | Organ |
| RI fenofibrate | Signal or action |
| FF induction | The effect of the signal on gene expression |
| RP P00543 | Number of regulatory unit that “contributes to/provides” this expression vector. |
| RS 2397 | Number of site that “contributes to/provides” this expression vector. |
| RR [Schoonjans K. et al., 1995] | Reference |
Fig. 2а. An example of description in the table TRRDEXP.
|
The fragment of the binding site description |
|
| AN 2397 | Number of site in TRRD |
| NM PPRE; peroxisome proliferator-response element | Short and full names of the site |
| … | |
| BF D38589:803 | Reference to EMBL |
Figure 2b. An example of description in the table TRRDSITES.
Description of transcription factors (table TRRDFAC)
Description of a transcription factor includes the species of the organism and the factor’s tissue or cellular location. In case the factor in question occurs in the cells under study in norm, it is indicated as “endogenous” in the corresponding field. The new format allows also the description of transcription factors of recombinant origin or synthesized in vitro. Illustrated in Fig. 2c is the description of the dimeric complex PPARalpha/RXRbeta, consisted of two in vitro synthesized subunits: Xenopus PPARalpha and mouse RXRbeta. This dimer was demonstrated to interact efficiently with the above-mentioned site AN 2397 in rat acyl-coenzyme A synthetase gene promoter resulting in transcription activation.
|
The description of the transcription factor that interacts with the corresponding binding site |
|
| TF PPARalpha/RXRbeta; PPARalpha and RXRbeta heterodimers | Short and full names of transcription factor |
| FS PPARalpha | Name of the described multimeric factor subunit |
| TS xenopus | Species to which the transcription factor belongs |
| NF T01352 | Number of the factor in the TRANSFAC database |
| TO in vitro synthesized | Origin of the factor |
| TR [Schoonjans K. et al., 1995] | Reference |
| FS RXRbeta | Name of the described multimeric factor subunit |
| … | |
Fig. 2c. An example of description in the table TRRDFAC.
Description of regulatory regions (TRRDGENES).
In addition to the description of the regulatory units (promoters, enhancers, silencers, etc.), the modified TRRD (Release 4.0) format allows the accumulation of the formalized data on location and characteristics of DNase I hypersensitive sites
Links to other databases and programs
The TRRD database comprises the five linked tables TRRDGENES, TRRDEXP, TRRDSITES, TRRDFAC, and TRRDBIB (Fig.1). In addition, the table TRRDGENES is furnished with the references to EMBL, SwissProt, EPD, EpoGERD, and EpoDB databases as well as to the GeneNet [Kolpakov, et al., 1998], which is a section of GeneExpress. The table TRRDSITES has the references to EMBL and TRANSFAC [Wingender et al., 1996] databases as well as to site recognition programs [Kondrakhin et al., in press] and the ACTIVITY database [Kolchanov et al., 1998b], included in the GeneExpress system [Kolchanov et al., 1998a]. The table TRRDFAC contains the references to TRANSFAC; the table TRRDBIB, to MEDLINE.
TRRD Viewer
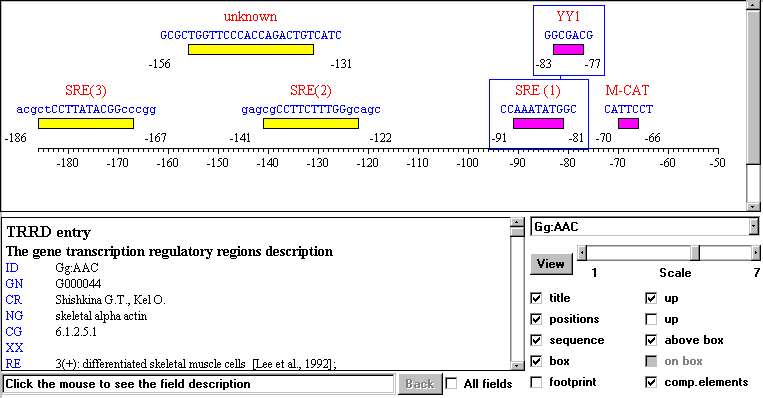
The applet TRRD-Viewer allows to visualize the data on the location of transcription factor binding sites in a map form (Fig. 3) and to overlook their textual description. When working with this applet, the user selects a gene identifier from the list, and the textual description of the gene (from TRRDGENES), its sites (from TRRDSITES), and the relevant references (from TRRDBIB) appear in the text window, while the transcription factor binding sites and composite elements are presented graphically. If the user clicks the site image, its description from the table TRRDSITES is displayed in the text window. Clicking the field title provides the display of the comments on the information described in the field. Options allowing different site representations are provided.
Fig. 3 An example of the visualization of the gene regulatory map by TRRD Viewer. Boxes represent binding sites for the transcription factors; line connecting rectangles indicates composite element
Contents of the TRRD database (Release 4.0)
The current release, TRRD 4.0, comprises the description of about 500 genes, over 800 regulatory units (promoters, enhancers, and silencers), and about 2400 transcription factor binding sites. Over 1500 scientific publications were processed to obtain these data. The genes described in TRRD belong to different eukaryotic species. Human (41%) and mouse (25%) genes constitute the major part; rat (15%) and chick genes are well represented too.
The TRRD database contains the information on genes encoding proteins with a wide variety of functions. F. Buher [Perier et al., 1998] suggested to classify the eukaryotic promoters basing on the function of the protein encoded. This classification reveals the following major gene groups in TRRD: genes for structural proteins (16%); genes for storage and transport proteins (20%); genes for enzymes (19%); genes for regulatory proteins, including hormones, growth factors, etc. (20%); and genes for proteins related to stress or pathogen defense reactions (10%). The rest 15% of the genes described have various other functions.
The TRRD database contains the sections describing transcription regulation of functionally significant gene groups (Table 1). Analysis of the data from these sections aidiid the clarification of the most important features of the transcription regulation of the gene groups in question [T.I. Merkulova et al., 1997; O.A. Podkolodnaya et al., 1997; E.A. Ananko et al., 1997; E.V. Ignateva et al., 1997; O.V. Kel and A.E. Kel, 1997].
| Name of section | Number of genes | Name and e.mail of the author |
| Interferon-Inducible Genes (IIG-TRRD) | 60 | Ananko E.A. eananko@bionet.nsc.ru |
| Erythroid-Specific Regulated Genes (ESRG-TRRD) | 44 | Podkolodnaya O.A. opodkol@bionet.nsc.ru |
| Genes of Lipid Metabolism (LM-TRRD) | 48 | Ignatieva E.V. eignat@bionet.nsc.ru |
| Glucocorticoid-Controlled Genes | 35 | Merkulova T.I. merk@cgi.nsk.su |
| Cell Cycle-Dependent Genes | 20 | Kel O.V. okel@ bionet.nsc.ru |
| Plant genes (Plant-TRRD) | 40 | Gorachkovskaya T.N. gocha@bionet.nsc.ru |
Table 1. The sections of the TRRD database
Conclusion
Thus, the modifications of the TRRD format used in TRRD Release 4.0 provide the user with a wider range of possibilities compared to the previous releases.The new scheme for description of gene expression patterns allows accumulation of the data in a formalized form and, what is most important, provides the possibility for their computer processing. The next task is to develop new mechanisms for the TRRD expanding. The work on the program for inputting data via the Internet, which would facilitate the TRRD development, is in progress [E. Ananko et al., 1998].
Acknowledgments
The work was supported by the Russian Foundation for Basic Research (96-04-50006, 97-04-49740, 98-04-49479), Integration Program of the Siberian Branch of the Russian Academy of Sciences (IGSBRAS-97*13), and Young Scientists Competition of SB RAS. The authors are grateful to I.V. LOKHOVA for assistance in bibliographic search and to D.VOROBIEV, D.KUROPATOV for creating links between TRRD and other computer systems and databases.
References
- A.E. Kel., N.A. Kolchanov, O.V. Kel, A.G. Romashchenko, E.A. Ananko, E.V. Ignatieva, T.I. Merkulova, O.A. Podkolodnaya, I.L. Stepanenko, A.V. Kochetov, F.A. Kolpakov, N.L. Podkolodnyi, and A.N. Naumochkin, “TRRD: Database on Transcription Regulatory Regions of Eukaryotic Genes” Mol. Biol. (Mosk.) 31, 521-530 (1997).
- O.V. Kel, “Structure of data representation in TRRD – database of transcription regulatory regions on eukaryotic genomes” Proceedings of the 28th Annual Hawaii International Conference on System Sciences [HICSS] 5. Biotechnology Computing, IEE Computer Society Press: Los Alamos, California 42 -51 (1995).
- F.A. Kolpakov, E.A. Ananko, G.B. Kolesov, and N.A. Kolchanov, “GeneNet: a database for gene networks and its automated visualization through the Internet” Bioinformatics, in press, (1998).
- N.A. Kolchanov, M.P. Ponomarenko, A.E. Kel, Y.V. Kondrakhin, A.S. Frolov, F.A. Kolpakov, O.V. Kel, E.A. Ananko, E.V. Ignatieva, O.A. Podkolodnaya, I.L. Stepanenko, T.I. Merkulova, V.N. Babenko, D.G. Vorobiev, S.V. Lavryushev, Y.V. Ponomarenko, A.V. Kochetov, G.B. Kolesov, N.L. Podkolodny, L. Milanesi, E. Wingender, T. Heinemeyer, and V.V. Solvyev “GeneExpress: A computer system for description, analysis, and recognition of regulatory sequences in eukaryotic genome” ISMB’98, in press (1998a)
- R.C. Perier, T. Junier, and P. Buher “The eukaryotic promoter database EPD” Nucleic Acids Res. 26, 353-357 (1998).
- N.A. Kolchanov, M.P. Ponomarenko, Yu.V. Ponomarenko, N.L. Podkolodny, and A.S. Frolov “Functional sites in the prokaryotic and eukaryotic genomes: computer models and activity predictions” Mol. Biol. (Mosk.) 32, ??? (1998b).
- Yu.V. Kondrakhin, V.N. Babenko, L. Milanesi, S.V. Lavryushev, and N.A. Kolchanov, “Recognition groups: a new method for description and prediction and of transcription factor binding sites” Comput. Appl. Biosci. (in press).
- E. Wingender, P. Dietze, H. Karas, and Kneuppel, “TRANSFAC: a database on transcription factors and their DNA binding sites” Nucleic Acids Res. 24, 238-241 (1996).
- E.V. Ignateva, T.I. Merkulova, O.V. Vishnevskii, and A.E. Kel, “Transcription regulation of lipid metabolism genes as described in the TRRD database” Mol. Biol. (Mosk.) 31, 575-591 (1997).
- T.I. Merkulova, V.M. Merkulov, and R.L. Mitina, “Glucocorticoid regulation mechanisms and glucocorticoid-controlled gene regulatory regions: description in the TRRD Database” Mol. Biol. (Mosk.) 31, 605-615 (1997).
- E.A. Ananko, S.I. Bazhan, O.E. Belova, and A.E. Kel, “Mechanisms of transcription regulation of interferon-inducible genes: description in the IIG-TRRD information system” Mol. Biol. (Mosk.) 31, 592-604 (1997).
- O.A. Podkolodnaya and I.L. Stepanenko “Mechanisms of transcription regulation of erythroid -specific genes” Mol. Biol. (Mosk.) 31, 562-574 (1997).
- O.V. Kel and A.E. Kel, “Complex gene network in cell cycle regulation: central role of the E2F family” Mol. Biol. (Mosk.) 31, 548-561 (1997).
- E.A. Ananko et al. This issue.