MERKULOVA T.I.+, MERKLOV V.M., KROPATCHEV K.Y., KALEDIN V.I.
Institute of Cytology and Genetics, (Siberian Branch of the Russian Academy of Sciences), 10 Lavrentieva ave., Novosibirsk, 630090 Russia
e-mail:merk@cgi.nsk.su
+Corresponding author
Keywords: glucorticoid-controlled gene, regilatory region, mechanism, hepatocarcirogens action, HNF3, DNA-binding activity, o-aminoazotoluene
The database GR-TRRD compiling the data on regulatory regions of 39 glucocorticoid-controlled genes is described. The database contains the description of over 80 glucocorticoid response elements (GREs) and 150 other regulatory elements, involved in tissue-specific, development and environmental regulation of glucocorticoid controlled genes. Comparative analysis of the regulatory regions of genes coding for tyrosine aminotransferase, aspartate aminotransferase and phosphoenolpyruvate carboxykinase allowed to propose that hepatocyte nuclear factor 3 (HNF3) is the likely target for hepatocarcinogen o-aminoazotoluene (ОАТ). This prediction has been confirmed by gel retardation experiments.
Introduction
Glucocorticoids influence a vast number of complex physiological and pathological processes. They help regulate protein, fat and carbohydrate metabolism, participate in immunosupression and anti-inflammation and are important for the optimum functioning of the cardiovascular, kidney, and nervous system. Glucocorticoids bring about their multiple effects by activating the intracellular glucocorticoid receptor (GR) that binds to specific glucocorticoid responsive elements (GREs) in the vicinity of regulated genes and subsequently affect their expression. For systematization, generalization, and analysis of constantly growing amount of experimental data on the glucocorticoid-controlled genes, the GR-TRRD (Glucocorticoid Regulated genes – Transcription Regulatory Regions Database) has been created as a constituent of the TRRD database [Kel’ A.E. et al., 1997]. The GR-TRRD is accessible at http://wwwicg.bionet.nsc.ru/trrd/34/gluc.htm.
Contents of the GR-TRRD database
The current release of the GR-TRRD (Release 4.0) compiles the data on 39 glucocorticoid-controlled genes (Table 1), including genes coding for enzymes of carbohydrate and amino acids metabolism (5), hormones(7) and hormone receptors(3), acute-phase proteins(7), serum(4), milk proteins(3), viral genes and others. Among them, 8 genes are of human origin; 18, rat; 4, mouse; 1, rabbit; 1, frog; 1, bovine; and 1, chicken. GR-TRRD contains the description of structural-functional organization of transcription regulatory regions of these genes, including the description of over 80 glucocorticoid response elements (GREs) and 150 other regulatory elements, involved in tissue-specific, development and environmental regulation of glucocorticoid controlled genes. An important component of GR-TRRD as a constituent of the TRRD database is RE field where the specificity of gene expression and regulation is described.
Transcription factor HNF3 is a target for hepatocarcinogen o-aminoazotoluene
Diminished glucocorticoid induction of tyrosine aminotransferase (ТАТ, G000798) gene expression is a well-known early effect of many hepatocarcinogens [Miller M.S.&Vogan G.N., 1986]. Meanwhile, it was shown that carcinogens do not reduce the glucocortiocoid inducibility of some other hepatic enzymes controlled by these hormones : aspartate aminotransferase (ASPART, G001175) and phosphoenolpyruvate carboxykinase(PEPCK, G000782) [Yeoh G.C.T, 1981 ]. So transcription factors that are involved in glucocorticoid regulation of the genes coding for TAT but not ASPART and PEPCK appear to be likely targets for carcinogen action. Comparative analysis of the regulatory regions of these genes described in the GR-TRRD allowed to propose that such are members of the HNF3 and Ets families [Merkulova T.I. et al., 1997] and GME-binding proteins [Oshima H. et al., 1995]. The most likely candidates are HNF3 family members, since TAT is expressed and regulated by glucocorticoids only in liver, while ASPART и PEPCK are also glucocorticoid induced in kidney, where HNF3 factors are virtually absent (G001244, G001010, G001242, G001172, G001240) (RE fields in corresponding entries of GR-TRRD).
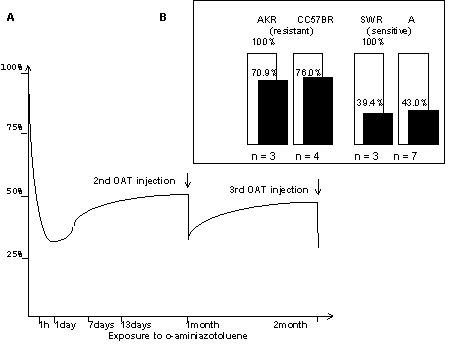
For that reason we assessed the DNA-binding activity of these factors in nuclear extracts from the livers of intact mice and those treated with the hepatocarcinogen o-aminoazotoluene (ОАТ). The experimental animal strains were ones with different sensitivity to liver tumor induction with OAT [Kaledin V.I.& Zakharova N.P.,1984]. DNA-binding activity of the AP1 family proteins, which can act, depending on the situation, either antagonistic to or synergistic with the glucocorticoid receptor [Merkulova T.I. et al., 1997], was measured as well. Gel retardation experiments demonstrated, that after OAT administration HNF3 DNA-binding activity was strongly reduced in the nuclear extracts from the livers of sensitive A and SWR mice, which have liver tumors induced with a high frequency (70-100%). Much less effect was found in the insensitive CC57Br and AKR strains, in which case induced tumours are rare (0-6%) (Fig.1A and the summary of the quantitative results in Figure 2B). The DNA-binding activity of Ets, AP1 factors and GME-binding proteins is not affected by OAT injection. The use of specific antisera against HNF3a, b and g allowed the HNF3 DNA-binding activity to be associated with the transcription factor HNF3 g ( Fig.1B). Treatment with OAT reduces the HNF3 DNA-binding activity within 1 hr after administration and keeps it so low during a months at least (Fig.2A)
Our data suggest that OAT affects the transcription factor HNF3g and therefore, the inhibitory effect, which this hepatocarcinogen has on the glucocorticoid induction of ТАТ, must be the result of reduction of its DNA-binding activity due to initiation by OAT of its direct or indirect modification, which is yet to be found out. As HNF3 is one of the most important liver-enriched transcription factors, responsible for differentiated liver cells phenotype, it suggests the involvement of the prolonged reduction of HNF3 DNA-binding activity in OAT hepatocarcinogenic action.
Acknowledgments
The work was supported by the Russian Foundation for Basic Research (98-04-49434)
Table 1. Contents of the GR-TRRD database
|
Functional groups |
Protein |
Gene N |
Regulation |
Species |
|
| I | Genes codingfor enzymes | Phosphoenolpyruvate(GTP)-carboxykinase | G000782 | positive |
rat |
| of carbohydrate and | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase | G000784 | positive |
rat |
|
| amino acids | Tyrosine aminotransferase | G000798 | positive |
rat |
|
| metabolism | Tryptophan oxygenase | G000803 | positive |
rat |
|
| Aspartate aminotransferase | G001175 | positive |
rat |
||
| II | Genes coding | Growth hormone | G000282 | positive |
human |
| for hormones | Placental lactogen 2 | G000235 | positive |
human |
|
| Prolactin | G001206 | negative |
cow |
||
| glycoprotein hormone a -subunit | G000271 | negative |
human |
||
| Pro-Opiomelanocortin | G000786 | negative |
rat |
||
| Angiotensinogen | G000703 | positive |
rat |
||
| Kininogen | G000797 | positive |
rat |
||
| III | Genes coding | Glucocorticoid receptor | G000742 | negative |
rat |
| for hormone | Insulin receptor | G000321 | positive |
human |
|
| receptors | Angiotensin receptor | G001204 | positive |
rat |
|
| IV | Viral genes | MMTV | G000455 | positive | |
| and | Murine Moloney Sarcoma | G001050 | positive | ||
| genomes | HPV-16 | G000171 | positive | ||
| V | Genes coding | a –acid glycoprotein | G000697 | positive |
rat |
| for acute | a –acid glycoprotein | G000463 | positive |
mouse |
|
| phase | Haptoglobin | G000751 | positive |
rat |
|
| proteins | Bb -fibrinogen | G001052 | positive |
frog |
|
| Interleukin-6 | G000316 | negative |
human |
||
| Metallothionein I | G000559 | positive |
mouse |
||
| Metallothionein IIА | G000337 | positive |
human |
||
| Gene coding for enzymes | Cytochrom CYP2B2 | G000729 | positive | ||
| of xenobiotic metabolism | Alcohol dehydrogenese 2 | G000185 | positive |
human |
|
| Genes coding for components of antibacterial defence | Lysozyme | G000064 | positive |
chiken |
|
| Genes coding for | b-casein | G000709 | positive |
rat |
|
| milk proteins | acid whay protein | G000629 | positive |
mouse |
|
| Genes | a2u-globulin | G000700 | positive |
rat |
|
| coding | a-fetoprotein | G000691 | negative |
rat |
|
| for serum | insulin-like growth factor binding protein 1 | G000310 | positive |
human |
|
| proteins | insulin-like growth factor binding protein 1 | G001207 | positive |
rat |
|
| Osteocalcin | G000353 | negative |
human |
||
| Osteocalcin | G000780 | negative |
rat |
||
| Proliferin | G000588 | negative |
mouse |
||
| Link protein | G001069 | positive |
rat |
||
| Uteroglobin | G000679 | positive |
rabbit |
Fig. 1. (A) AP1, GME , HNF3 and Ets DNA-binding activity. Every set of data represents: left – the free binding site (at the bottom of the gel), middle – binding site after incubation with nuclear extracts from the livers of intact mice, right – mice treated with the hepatothropic carcinogen OAТ.
(B) Identification of HNF3 isoforms that bind to HNF3 element (A mice). C-control
Fig 2.(A) Time course for HNF 3-DNA binding activity in response to OAT at A mice
(B) Changes in HNF 3-DNA binding activity 24 hours after one-time OAT administration
References
- A.E. Kel., N.A. Kolchanov, O.V. Kel, A.G. Romashchenko, E.A. Ananko, E.V. Ignateva, T.I. Merkulova, O.A. Podkolodnaya, I.L. Stepanenko, A.V. Kochetov, F.A. Kolpakov, N.L. Podkolodnyi, and A.N. Naumochkin, “TRRD: database on transcription regulatory regions of eukaryotic genes” Mol. Biol. 31, 521-530 (1997).
- V.I. Kaledin and N.P. Zakharova, In: Investigation on tumor induction and metastasizing in experimental animals. Novosibirsk, 146-185, (1984).
- T.I. Merkulova, V.M. Merkulov and R.L. Mitina “Glucocorticoid mechanisms and glucocorticoid controlled gene regulatory regions: Description in the TRRD database” Mol. Biol. 31, 714-726 (1997).
- M.S. Miller and G.N. Vogan, “Inhibition of steroid-inducible tyrosine aminotransferase gene expression by N-methyl-N’-nitrosoguanidine in a rat hepatoma cell line”. Carcinogenesis 7, 1273-1278 (1986).
- H. Oshima, D. Szapary and S.S. Simons ”The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquites heteromeric complex”.J.Biol.Chem. 270, 21893-21901(1995).
- G.C.T. Yeoh, “The effect of 3’-methyl-4-dimethyl-aminoazobenzene on foetal rat hepatocytes in culture” Eur. J. Cancer Clin. Oncol. 17, 743-752 (1981)