Laboratory of Theoretical Genetics, Institute of Cytology and Genetics (Siberian Branch of the Russian Academy of Sciences),
Lavrentieva 10, Novosibirsk, 630090 Russia;
+Corresponding author e-mail:eignat@bionet.nsc.ru
Keywords: lipid metabolism, transcription, regilatory regions database, transcription factor, genes, binding sites, gene networks, mevalonate pathway, cholesterol, biosyntesis
The database LM-TRRD compiling the data on genes of lipid metabolism (LM) is described. Over 30 different transcription factors are involved in the transcription regulation of the lipid metabolism genes. Among them, transcription factors SREBP, PPAR, HNF-4, RXR, and COUP-TF are most important for LM regulation. The transcription factors belonging to the PPAR and SREBP subfamilies are the key elements providing the functioning of two LM subsystems: fatty acid oxidation (PPAR factors) and cholesterol homeostasis control (SREBP factors).
Introduction
Lipids comprise one of the most important classes of complex molecules present in animal cells and tissues. Lipid diversity and level in the cells, tissues, and organs are determined by the processes of lipid metabolism (LM), which include lipid transport, consumption and intracellular utilization, de novo synthesis, degradation, and excretion. The processes of lipid metabolism require the involvement of numerous proteins with different functions. These proteins together with their genes are the components of the LM system. The interest for the LM system is due to its important role in the vital activity of the organism and to the fact that the distortions in its functioning are among the causes of different human diseases. The amount of experimental data on different peculiarities of functioning of this system has grown enormous during the past years including those on the transcription regulation of the genes of lipid metabolism. For systematization, generalization, and analysis of data on lipid metabolism system, the LM-TRRD (Lipid Metabolism – Transcription Regulatory Regions Database) has been created as a constituent of the TRRD database. The current release of the LM-TRRD compiles the data on the genes of the LM system in TRRD format [N.A. Kolchanov, 1998, this issue] [Kel’ A.E. et al., 1997]. The LM-TRRD is accessible at http://wwwicg.bionet.nsc.ru/trrd/34/lipmet.htm. In addition, since LM-TRRD is a section of the TRRD, the information on transcription regulation of the LM system genes can be obtained through the SRS (http://wwwmgs.bionet.nsc.ru/), which performs the search within the entire TRRD database [N.A. Kolchanov, 1998, in press].
Contents of the LM-TRRD database
The current release of the LM-TRRD (Release 4.0) compiles the data on 48 genes of the lipid metabolism system (Table 1): genes of enzymes involved in lipid biosynthesis and degradation (16), genes of transport proteins (18), genes of cell surface receptors (3), genes of hormones and genes of transcription factors (9). Among them, 21 genes are of human origin; 7, mouse; 15, rat; 1, rabbit; 1, frog; 2, hamster; and 1, chicken. The database contains the description of over 60 regulatory regions (promoters, enhancers, and silencers) of lipid metabolism genes. Note that a number of regulatory regions is distanced from the transcription start by several kbp, for example the enhancer for mouse HNF-4 gene (G001173) is located 7 kbp in the 5’ direction from the transcription start; the enhancer for rat hydratase-dehydrogenase gene (G000757) is located 3 kbp upstream of the transcription initiation site. Most genes are transcribed from one promoter; however, certain genes (G000684, G001191, and G001195) have alternative promoters providing the transcription either in different tissues or under the effect of different inducers. Over 300 various transcription factor binding sites occurring in the regulatory regions of LM genes are described in the LM-TRRD.
Table 1. Contents of the LM-TRRD database
| Gene |
Gene number in TRRD (spicies) |
Transcription factors regulating gene expression |
|
I. Genes of the transport proteins |
||
| apolipoprotein(a) | G000208(human) | HNF-1, C/EBP, HNF-4 |
| apolipoprotein A-I | G000203(human) | HNF-3, HNF-4, C/EBP, RAR/RXR, ARP-1, Ear3/COUP-TF, PPAR, Egr-1, Sp-1 |
| G000705(rat) | HNF-4, ARP-1 | |
| apolipoprotein AII | G000204(human) | HNF-1, HNF-4, C/EBP, ARP-1, Ear3/COUP-TF, NF-BA1, CIIIB1, AP-1, TF-LF1, NF1 |
| apolipoprotein A-IV | G001066(human) | HNF-4, Arp-1, Ear3/COUP-TF |
| apolipoprotein B | G000205(human) | HNF-1, HNF-3, HNF-4, C/EBP, ARP-1, Ear3/COUP-TF, NF-BA1, AF-1 |
| apolipoprotein C-III | G000206(human) | HNF-4, PPAR, ARP-1, Ear3/COUP-TF, NF-BA1, NF-k B, CIIIB1 |
| G000706(rat) | HNF-4, PPAR/RXRalpha | |
| apolipoprotein E | G000207(human) | Sp-1, BEF-1 |
| apoVLDL II | G000048(chicken) | VBP, ER, C/EBP, COUP-TF |
| serum amyloid A2 | G000382(human) | C/EBPb , NF-k B |
| serum amyloid Ag9 | G000384(human) | NF-k B |
| serum amyloid A3 | G000599(mouse) | C/EBPb , NF-k B, SEF-1 |
| serum amyloid A | G000678(rabbit) | C/EBPb , NF-k B, SAF |
| serum amyloid A1 | G000794(rat) | C/EBPb , NF-k B, YY1 |
| cellular retinol-binding | G000727(rat) | RAR/RXR |
| protein II | G001210(mouse) | HNF-4, Arp-1, RAR/RXR |
| intestinal fatty acid binding protein | G001199(rat) | HNF-4, ARP-1 |
|
II Genes of the cell surface receptors |
||
| macrophage scavenger receptor gene | G000387(human) | Ets, AP-1, PU.1/Spi-1 |
| alpha2-macroglobulin receptor/lipoprotein receptor-related protein gene | G001200(human) | Sp1 |
| LDL receptor | G001190(human) | SREBP, Sp-1 |
|
III. Genes of the enzymes involved in lipid biosynthesis |
||
| acetyl-CoA carboxylase | G000684(rat) | AP-2, SREBP-1, Sp-1, C/EBPb |
| fatty acid synthase | G001196(rat) | SREBP-1, Sp1 |
| farnesyl diphosphate synthase | G001191(rat) | SREBP-1, NF-Y |
| mitochondrial glycerol-3-phosphate acyltransferase gene | G001197(mouse) | |
| 3-hydroxy-3-methylglutaryl-CoA synthase | G001198 (hamster) | AP-1, SREBP-1, NF-Y |
| 3-hydroxy-3-methylglutaryl-CoA reductase | G000157(hamster) | SREBP, Red25 |
| squalene synthase | G001183(human) | ADD1/SREBP-1 |
|
IV. Genes of the enzymes involved in lipid degradation |
||
| hepatic triglyceride lipase | G000288(human) | HNF-1, C/EBP |
| acyl-coenzyme A synthetase | G001195(rat) | PPAR/RXR |
| lipoprotein lipase | G000329(human) | |
| medium-chain acyl coenzyme A dehydrogenase | G001201(human) | HNF-4, RAR/RXR, Ear3/COUP-TF, Sp1 |
| hydratase-dehydrogenase | G000757(rat) | PPAR/RXR |
| acyl-CoA oxidase | G000704(rat) | PPAR |
| G001209(human) | PPAR/RXR | |
| cholesterol 7alpha- | G001193(human) | BARP |
| hydroxylase (CYP7) | G001194(rat) | BARP |
|
V. Genes of the hormones |
||
| leptin | G001032(mouse) | C/EBPa , C/EBPb |
| leptin | G001258(human) | C/EBPa |
|
VI. Genes of the transcription factors involved in the regulation of lipid metabolism system |
||
| C/EBPa | G000490(mouse) | C/EBPa , NF-1, USF, Myc/Max, CUP, Sp1 |
| G001170(human) | USF, Sp1 | |
| G001192(rat) | C/EBPa | |
| HNF-1 | G000756(rat) | HNF-1, HNF-4 |
| G001171(xenopus) | HNF-1, HNF-4 | |
| HNF-3b | G001172(rat) | HNF-3b , VBP, C/EBPb |
| HNF-4 | G001173(mouse) | HNF-1a |
| RARb | G000376(human) | ATF/CREB, RAR |
| Egr-1 | G000505(mouse) | Ets, SRF |
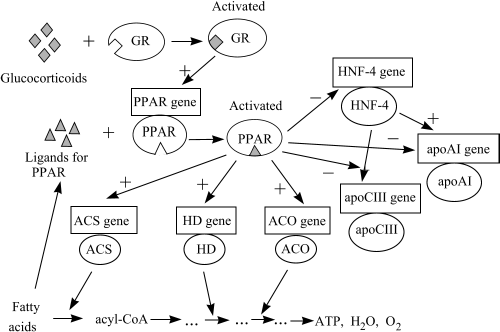
Fig. 1. Gene network in the system of genes regulated by PPAR factors. Here and in Fig. 2, circles indicate proteins; rectangles show genes coding for these proteins. ACO, acyl-CoA oxidase; ACS, acyl-coenzyme A synthetase; poAI – apolipoprotein AI; apoCIII – apolipoprotein CIII; GR – glucocorticoid receptor; HD, Hydratase-dehydrogenase; HNF4, hepatocyte nuclear factor 4; PPAR – peroxisome proliferator activated receptor;
Analysis of the data contained in the LM-TRRD have demonstrated that over 30 transcription factors are involved in the transcription regulation of the LM genes [Ignati’eva et al., 1997]. These factors differ in the structure of their DNA-binding domain and belong to different families.
Transcription factors SREBP, PPAR, HNF-4, RXR, and COUP-TF play a particular role in transcription regulation of LM genes. The binding sites of these factors are nonrandomly frequent in the regulatory regions of the LM genes compared with the other genes described in the TRRD [Ignati’eva et al., 1997]. Note that four (PPAR, HNF-4, COUP-TF, and RXR) of the five factors mentioned above contain the DNA-binding domain of “zinc finger” type. In addition, the factors belonging the COUP-TF family may be considered as major negative regulators of LM gene transcription, as 13 cases of transcription inhibition of the 16 described in the LM-TRRD database is caused by the factors of this family.
Gene networks
Based on the LM-TRRD-contained information on transcription regulation of various vertebrate genes, we have designed two fragments of the lipid metabolism gene network. The first fragment describes coordination of the genes controlling the fatty acid oxidation (Fig. 1). Involved in this process are the following genes of enzymes of fatty acid oxidation and proteins for lipid transport: ACS (G001195), HD (G000757), ACO (G000704) and apoAI (G000203), apoCIII (G000206), respectively. The PPAR transcription factors form the key component of this gene network. Glucocorticoids are known to activate the expression of the gene encoding one of the PPAR factors in liver [Lemberger T. et al., 1994]. In addition, the interaction with fatty acids or their derivatives increases the activity of PPAR subfamily proteins [Schoonjans K. et al., 1995]. Thus, the PPAR subfamily factors provide for the expression regulation of the above-listed genes of enzymes and transport proteins depending on the presence of fatty acids in the cell and in response to glucocorticoids.
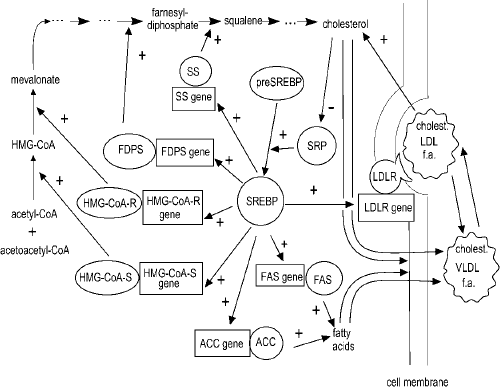
Fig.2. Gene network of a subsystem of intracellular cholesterol control. ACC, acetyl coenzyme A carboxylase; f.a. – fatty acids; FAS, fatty acid synthase; FDPS – farnesyl diphosphate synthase; HMG-CoA-R, 3-hydroxy-3-methylglutaryl CoA reductase; HMG-CoA-S, 3-hydroxy-3-methylglutaryl CoA synthase; LDL, low density lipoprotein; LDLR, low density lipoprotein receptor; preSREBP sterol regulatory element-1 binding protein precursor; SREBP – sterol regulatory element-1 binding protein; SRP – sterol-regulated protease; SS, squalene synthase; LDL, very low density lipoprotein
The second gene network fragment unites the genes controlling the cholesterol level in the cell (Fig. 2). This fragment includes the genes of enzymes involved in mevalonate pathway of cholesterol biosynthesis and fatty acid biosynthesis: HMG-CoA-S (G001198), HMG-CoA-R(G000157), FDPS (G001191), SS (G001183) and ACC (G000684), FAS (G001196), respectively, as well as LDLR (G00190). Transcription factors of SREBP subfamily are the key elements of this gene network. It has been demonstrated that SREBP transcription factors are formed in the cell from a precursor molecule (preSREBP) by sterol-regulated protease (SRP) [X. Wang et al., 1994]. SRP activity is, in turn, inhibited by a high cholesterol content in the cell [X. Wang et al., 1994, X. Wang et al., 1995]. Thus, SREBP proteins are involved in transcription regulation of the group of genes in question and provide for either their coordinated activation or expression inhibition depending on the cholesterol level in the cell.
Conclusion
Peculiarities of the lipid metabolism gene system regulation have been studied on the example of 48 genes contained in the LM-TRRD database. The set of transcription factors involved in this regulation has been determined. The accumulated information on primary sequences of the binding sites of these transcription factors is of great value. The nearest goal is to expand the range of the genes described in the LM-TRRD database. In the next releases of the database, special attention will be paid to the genes expressed in fat tissue as well as the genes responsible for hormonal regulation of lipid metabolism processes.
Acknowledgments
The work was supported by the Russian Foundation for Basic Research (98-04-49479). The author is grateful to N.A. KOLCHANOV for scientific guidance; to T.I. MERKULOVA, O.V. KEL, and A.E. KEL for valuable discussions and advice; to E.A. ANANKO, O.A. PODKOLODNAYA, F.A. KOLPAKOV, S.V. LAVRYUSHEV, D.A. GRIGOROVICH, D.VOROBIEV, N.L. PODKOLODNY, A.S. FROLOV for their help in connecting the database with the Internet; and I.V. LOKHOVA for assistance in bibliographic search.
References
- A.E. Kel., N.A. Kolchanov, O.V. Kel, A.G. Romashchenko, E.A. Anan’ko, E.V. Ignateva, T.I. Merkulova, O.A. Podkolodnaya, I.L. Stepanenko, A.V. Kochetov, F.A. Kolpakov, N.L. Podkolodnyi, and A.N. Naumochkin, “TRRD: database on transcription regulatory regions of eukaryotic genes” Mol. Biol. 31, 521-530 (1997).
- N.A. Kolchanov, E.V. Ignatieva, O.V. Kel-Margoulis, A.E. Kel, E.A. Ananko., O.A. Podkolodnaya, et al., “Transcription regulatory regions database (TRRD): new possibilities provided by RELEASE 4.0” This issue (1998)
- N.A. Kolchanov, M.P. Ponomarenko, A.E. Kel, Y.V. Kondrakhin, A.S. Frolov, F.A. Kolpakov, O.V. Kel, E.A. Ananko, E.V. Ignatieva, O.A. Podkolodnaya, I.L. Stepanenko, T.I. Merkulova, V.N. Babenko, D.G. Vorobiev, S.V. Lavryushev, Y.V. Ponomarenko, A.V. Kochetov, G.B. Kolesov, N.L. Podkolodny, L. Milanesi, E. Wingender, T. Heinemeyer, and V.V. Solovyev “GeneExpress: a computer system for description, analysis, and recognition of regulatory sequences in eukaryotic genome” ISMB’98, in press
- E.V. Ignatieva, T.I. Merkulova, O.V. Vishnevskii, and A.E. Kel “Transcription regulation of lipid metabolism genes as described in the TRRD database” Mol. Biol. 31, 575-591 (1997)
- T. Lemberger, B. Staels, R. Saladin, B. Desvergne, J. Auwerx, and W. Wahli “Regulation of the peroxisome proliferator-activated receptor alpha gene by glucocorticoids” The Journal of Biological Chemistry 269, 40, 24527-24530 (1994)
- K. Schoonjans, M. Watabene, H. Suzuki, A. Mahfoudi, G. Krey, W. Wahli, P. Grimaldi, B. Staels, T. Yamamoto, and J. Auwerx “Induction of the acyl-coenzyme A synthase gene by fibrates and fatty acids is mediated by peroxisome proliferator response element in the C promoter” The Journal of Biological Chemistry 270, 33, 19269-19276 (1995)
- X. Wang, R. Sato, M. Brown, X. Hua, and J.L. Goldstein “SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis” Cell 77, 9, 53-62 (1994)
- X. Wang, J.-T. Pai, E.A. Wiedenfeld, J.C. Medina, C.A. Slaughter, J.L. Goldstein, and M.S. Brown “Purification of an interleukin-1 beta converting enzyme-related cysteine protease that cleaves sterol regulatory element-binding proteins between the leucine zipper and transmembrane domains” The Journal of Biological Chemistry, 270, 30, 18044-18050 (1995)