GORYACHKOVSKY T.N., ANANKO E.A., PELTEK S.E.
Institute of Cytology and Genetics, (Siberian Branch of the Russian Academy of Sciences), 10 Lavrentieva ave., Novosibirsk, 630090 Russia
Keywords: plant, database, expression regulation, transcription, DNA regulatory regions, genes, storage protein genes, genes of photosyntesis, nitrogen fixation, transcription factor, binding sites
A large volume of information on the structure and expression regulation of various plant genes has been accumulated [Neto et al., 1995; Ellerstrom et al., 1996; Gallusci et al., 1994; Howley & Gatehouse, 1997; Nakase et al., 1997].
The key events of expression regulation of any genes occur at the transcription level. Interaction of protein factors with DNA regulatory regions determines the time, place, and expression level of the gene under control. Ontogenesis of each organism implies the consequent ordered switching the genes on and off and modulation of their. Certain complexly organized transcription systems have been formed in higher plants. To generalize, analyze, and systematize these data, the PLANT-TRRD database is being created as a constituent of the TRRD (Transcription Regulatory Regions Database) [A.E. Kel et al., 1997].
The TRRD database is integrated into the Interment-accessible GeneExpress system ( http://wwwmgs.bionet.nsc.ru/systems/GeneExpress/).
The TRRD format meets the up-to-date concept of the structure and functioning of eukaryotic gene regulatory regions. Continuous DNA regions containing regulatory elements may occur in 5’-, 3’-flanking regions, or introns, thereby forming regions of transcription regulation. The regions of transcription regulation provide for gene-specific transcription complex formation. These complexes determine the transcription rate depending on the ontogenetic stage, organ, environmental conditions, etc. Transcription regulation of individual genes is coordinated within gene networks [Ananko et al., 1997; Ignat’eva et al., 1997; Kolpakov et al 1998]. Coordinated expression of such gene groups is important for performance of particular functions of cells, tissues, organs, regulation of ontogenesis, and response to environmental conditions. In addition to the transcription level, the regulation within the frames of gene networks occurs at the levels of translation, splicing, post-translational processing of proteins, and several other processes. Interactive regulation of the gene network elements is performed through transcription and membrane factors, hormones, and enzymes. The current release of PLANT-TRRD contains the data on regulatory regions of 38 plant genes (Table 1). Genes of storage proteins of plant seeds constitute the major part of the PLANT-TRRD database. Storage proteins fulfill the same function in various species, namely, supply the developing embryo with nitrogen, sulfur, and carbon; their genes are regulated by similar mechanisms and regulatory elements. Expression of storage protein genes is regulated at both transcriptional and post-transcriptional levels [Ellerstrom et al., 1996; Albani et al., 1997].
Realization of an individual function requires, as a rule, a synchronized activation of a group of genes. In certain cases, it is performed at the level of transcription regulation. For example, the genes expressed in the endosperm contain a conservative element, the so-called endosperm box, in their promoter in the region of – 300 bp from the transcription start [Neto et al., 1995; Hammond-Kasack et al., 1993].
In addition to storage protein genes, the database contains the sections devoted to genes of photosynthesis and nitrogen fixation systems, genes of enzymes, plant viruses, etc. Each gene included in the TRRD database is represented by a separate entry containing general information on the gene and its regulatory regions. Description of transcription regulatory regions includes gene characterization and links to other databases containing additional information on regulation peculiarities of this gene (TRANSFAC, COMPEL, and EPD) [Heinemeyer et al., 1998; Perier et al., 1998].
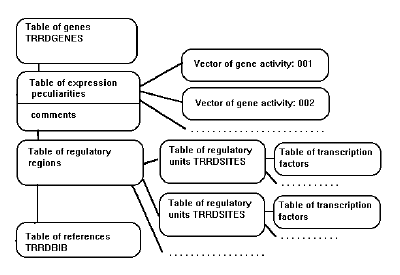
These fields in each card are grouped in tables (Fig. 1). Table of genes (TRRDGENES) contains entry identifier, species and gene names, date of last modification, key words, and chromosomal location if available.
Table 1. PLANT-TRRD content.
| Gene | Organism | Region |
| Monocot seed storage proteins | ||
| LMW glutenin 1D1 | wheat, Triticum aestivum | ST: -300 to +30 |
| HMW glutenin D1-2 | wheat, Triticum aestivum | ST: -375 to -45 |
| alpha-zein | maize, Zea mays | ST: -362 to 317 |
| beta-zein | maize, Zea mays | SR: -233 to +81 |
| gamma-zein | maize, Zea mays | ST: -1024 to +61 |
| alpha-coixin | Job’s tears, Coix lacryma-jobi | SR: -295 to +45 |
| B-coixin | Job’s tears, Coix lacryma-jobi | SR: -233 to -13 |
| C-hordein | barley, Hordeum vulgare | ST: -431 to -126 |
| glutelin A-1 | rice, Oryza sativa | ST: -595 to -86 |
| glutelin A-2 | rice, Oryza sativa | ST: -5100 to +1 |
| glutelin A-3 | rice, Oryza sativa | ST: -439 to -316 |
| glutelin B-1 | rice, Oryza sativa | ST: -621 to +18 |
| 16 kDa albumin G1 | rice, Oryza sativa | ST: -557 to -152 |
| alpha-globulin-1 | rice, Oryza sativa | ST: -754 to -89 |
| prolamin 5a | rice, Oryza sativa | ST: -642 to -155 |
| Dicot seed storage proteins genes | ||
| helianthinin G3D | sunflower, Helianthus annuus | ST: -875 to +1 |
| helianthinin G3A | sunflower, Helianthus annuus | ST: -1700 to +1 |
| legumin A | pea, Pisum sativum | ST: -1204 to +45 |
| beta-phaseolin | french bean, Phaseolus vulgaris | ST: -422 to -13 |
| b -conglycinin a ‘-subunit | soybean, Glycine max | ST: -257 to +14 |
| napin A | oilseed rape, Brassica napus | ST:-309 to +44 |
| Low-temperature regulated proteins | ||
| ADH-1 | maize, Zea mays | ST: -260 to +1 |
| BN115 | oilseed rape, Brassica napus | ST: -300 to -25 |
| Heat shock proteins | ||
| Gmhsp 18.5-C | soybean, Glycine max | SR: -900 to +730 |
| Gmhsp 17.9-D | soybean, Glycine max | SR: -1264 to +1163 |
| Genes of photosynthesis system | ||
| rbcS-1A | mouse-ear cress, Arabidopsis thaliana | ST:-359 to -278 |
| rbcS 3A | tomato, Lycopersicon esculentum | ST: -359 to -278 |
| PEPC | maize, Zea mays | ST: -550 to -30 |
| phytochrome A | rice, Oryza sativa | ST: -364 to +1 |
| Embryo-specific genes | ||
| Em | wheat, Triticum aestivum | ST: -168 to +1 |
| Abscisic acid-responsive genes | ||
| rab-16A | rice, Oryza sativa | ST: -294 to -52 |
| Transcription factors | ||
| Opaque 2 | maize, Zea mays | ST: -190 to +1 |
| Flavonoid glucoside pathway genes | ||
| chalcone synthase | petunia, Petunia hybrida | ST: -2400 to +81 |
| chalcone synthase | snapdragon, Antirrhinum majus | ST: -357 to -39 |
| Nodul-specific genes | ||
| nodulin N23 | soybean, Glycine max | ST: -392 to +1 |
| leghaemoglobin Lbc3 | Soybean, Glycine max | ST: -1100 to -49 |
| Plant viruses | ||
| CaMV | Cauliflower mosaic virus | ST: -90 to +2 |
| MSV coat protein | Maize Streak Virus | GS: 2450 to 2570 |
This block includes the information on the relative location of regulatory regions and various starting points used by different authors. Information on alignment of long regulatory regions of various genes is provided as well as the comments on the entire gene and interactions of various regulatory regions.
Figure 1. Structure of PLANT-TRRD database.
References to GenBank, EMBL, or SWISS-PROT are given. The next block describes the peculiarities of the gene expression. The fields of this block contain the data on time and place of expression. A separate block of fields, vector of gene activity, in each card is devoted to every instance of expression described. The fields of the next block describe the entire regulatory region and particular regulatory regions that constitute it, and individual transcription factor binding sites. Having chosen a site of interest, user will get its detailed description in the following fields:
Table 2. Regulatory unit
| A fragment of the card on beta-zein G001259 | Content of informational field |
| AN 2457 | Number of the site in the database |
| NM -300 element; | Name of the site |
| SQ tatcgttaCACATGTGTAAAGGT | Sequence |
| PQ -331 to –317 | Positions |
| PF -339 to –318 | Footprint positions |
| BF V01472:119 | Reference to EMBL |
| AG endosperm tissues : 1.1.1, 3.1, 9 | Name of cell line and experiment code |
| HM This element contains part of … | Homologous site |
| KK The protein binding… | Comments on the site |
The description of a regulatory site is linked to the block describing the transcription factor interacting with this site (Fig. 1).
The fields of TRRDBIB contain the information on the relevant publications and reference to MEDLINE.
Several key events take place in the course of plant ontogenesis; the first of them is seed germination. As a rule, this process is under environmental control. Humidity, temperature, light regime, etc., form the optimal conditions triggering the ontogenesis per se, that is, a successive switching definite genes on and off. The other type of genes provides for morphogenesis. A part of these genes are regulated by inner mechanisms alone, while expression of the others are modulated by environmental conditions. The genes responsible for interaction of the organism with environment are of prime importance too. Belonging to this group are the genes induced by stress, low temperature or heat shock, and various chemical substances. The genes expressed throughout the entire life of the plant provide for the vital physiological needs and plant growth. These are housekeeping genes, genes encoding enzymes and hormones, genes providing secretion of various substances, etc.
The PLANT-TRRD database will be further developed basing on functional grouping of the genes into gene networks. This means that each section of the database will describe the genes providing for a certain function, such as, for example, storage proteins, heat shock genes, genes of photosynthesis system, etc.
Acknowledgments
The work was supported by the Russian Foundation for Basic Research (98-04-49479). The author is grateful to N.A. KOLCHANOV for scientific guidance; to O.A. PODKOLODNAYA, F.A. KOLPAKOV, S.V. LAVRYUSHEV, D.A. GRIGOROVICH, D.VOROBIEV, N.L. PODKOLODNY, A.S. FROLOV for their help in connecting the database with the Internet; and I.V. LOKHOVA for assistance in bibliographic search.
References
- D. Albani, M.C.U. Hammond-Kosack, C. Smith, S. Conlan., V. Colot, M. Holdsworth, and M.W. Bevan, “The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes” Plant Cell 9, 171 (1997).
- E.A. Ananko, S.I. Bazhan, O.E. Belova, and A.E. Kel, “Mechanisms of transcription regulation of interferon-inducible genes: description in the IIG-TRRD information system” Mol. Biol. (Msk.) 31, 701 (1997).
- R.C. Perier, T. Junier and P. Bucher, “The eukaryotic promoter database EPD” Nucleic Acids Res. 26, 353 (1998).
- M. Ellerstrom, K. Stalberg, I. Ezcurra, and L Rask, “Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription” Plant Mol. Biol. 32, 1019 (1996).
- P. Gallusci, F. Salamini and R.D. Thompson, “Differences in cell type-specific expression of the gene Opaque 2 in maize and transgenic tobacco” Mol. Gen. Genet. 244, 391 (1994).
- M.C.U. Hammond-Kasack, M.J. Holdsworth and M.W. Bevan, “In vivo footprinting of a low molecular weight glutenin TI gene (LMWG-1D1) in wheat endosperm” EMBO J. 12, 545 (1993).
- T. Heinemeyer, E. Wingender, I. Reuter, H. Hermjakob, A.E. Kel, O.V. Kel, E.V. Ignatieva, E.A. Ananko, O.A. Podkolodnaya, F.A. Kolpakov, N.L. Podkolodny and N.A. Kolchanov, “Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL” Nucleic Acids Res. 26, 362
- P.M. Howley and J.A. Gatehouse, “A 38 bp repeat sequence within the pea seed storage protein promoter of legA is a binding site for nuclear DNA-binding protein” Plant Molecular Biology 33, 175 (1997).
- E.V. Ignateva, T.I. Merkulova, O.V. Vishnevskii, and A.E. Kel, “Transcription regulation of lipid metabolism genes as described in the TRRD database” Mol. Biol. (Msk.) 31, 684 (1997).
- A.E. Kel, N.A. Kolchanov, O.V. Kel’, A.G. Romashchenko, A.G. Ananko, E.V. Ignateva,
- T.I. Merkulova, O.A. Podkolodnaya, I.L. Stepanenko, A.V. Kochetov, F.A. Kolpakov, N.L. Podkolodnyi, and A.A. Naumochkin, “TRRD: database on transcription regulatory regions of eukaryotic genes” Mol. Biol. (Msk.), 31, 626 (1997).
- F.A. Kolpakov, E A. Ananko, G.B. Kolesov and N.A. Kolchanov, “GeneNet: a database for gene networks and its automated visualization through the Internet” in press (1998).
- G.C. Neto, J.A. Yunes, M.J. da Silva, A.L. Vettore, P. Arruda, and A. Leite, “The involvement of Opaque 2 on beta-prolamin gene regulation in maize and Coix suggests a more general role for this transcriptional activator” Plant Mol. Biol. 27, 1015 (1995).
- M. Nakase, T. Yamada, T. Kira, J. Yamaguchi, N. Aoki, R. Nakamura, T. Matsuda, and T. Adachi, “The same nuclear proteins bind to the 5′-flanking regions of genes for the rice seed storage protein: 16 kDa albumin, 13 kDa prolamin and type II glutenin: Plant Mol. Biol. 32, 621 (1996).