KEL-MARGOULIS O.V.+, KEL A.E., FRISCH M.#, ROMASHCHENKO A.G., KOLCHANOV N.A., WINGENDER E.#
Institute of Cytology and Genetics, (Siberian Branch of the Russian Academy of Sciences), 10 Lavrentieva ave., Novosibirsk, 630090 Russia, okel@bionet.nsc.ru;
#Gesellschaft fur Biotechnologische Forschung mbH, Mascheroder Weg 1, D-38124 Braunschweig, Germany, ewi@gbf.de
+Corresponding author
Keywords: database, composite regulatory elements, DNA-protein interaction, protein-protein interaction, gene-specific regulation, binding sites, synergism or antagonism, transcription factor, tissue-specific induction, inducible regulation, DNA-binding domains
The database COMPEL collects information on composite regulatory elements, which are minimal functional units providing combinatorial transcriptional regulation due to specific protein-DNA and protein-protein interactions. There are composite elements of synergistic and antagonistic type depending on the character of interactions between the transcription factors involved. Composite elements can be classified based on the type of factors‘ DNA-binding domain as well as on regulatory pattern provided by the composite element. In the present paper we describe a suggested relational model of the database and its integration with TRANSFAC and TRRD databases. Presently the database contains about 200 composite elements and is electronically available under SRS: http://transfac.gbf.de/srs5/ and by FTP: ftp://transfac.gbf.de/pub/databases/compel/ or ftp://ftp.bionet.nsc.ru/pub/biology/compel/.
1. Introduction
Protein-protein interactions play a major role in gene-specific regulation of transcription. They occur between general transcription factors and RNA polymerase II in the course of basal transcription complex formation, between activation domains of upstream binding factors and certain general factors, and between transcription factors binding to closely situated target sites as well. We will refer to a pair of two closely situated binding sites that acquire new regulatory properties due to direct or indirect interactions between corresponding transcription factors as a composite element (CE) [1-3]. Structurally similar elements are present in several different genes, which apparently implies that such regulatory modules are functionally significant. For instance, composite elements containing binding sites for SRF and Ets family factors were found in three early response genes, c-fos (human), egr-1 (mouse) and pip92 (mouse) (COMPEL acc numbers C00022, C00126, C00127). The CEs that include NF-k B and C/EBP-b binding sites have been detected in eight genes highly inducible upon acute-phase response (COMPEL acc numbers C00098, C00100, C00101, C00152-C00155).
There are two main types of composite elements: synergistic and antagonistic ones. In synergistic CEs simultaneous interactions of two factors with closely situated target sites result in a high level of transcriptional activation. Highly cooperative binding of factors to DNA and formation of a ternary complex protein-protein-DNA was experimentally shown in many cases. As a result of protein-protein interactions a new protein surface may be formed which is common for factor pair. In some cases two factors independently binding to DNA, still synergistically activate transcription. In this case synergistic effect may be accounted for by interactions of these factors with different general factors of a basal transcription complex. Some factors are known to bend DNA and thus permit binding of other factors. Within an antagonistic CE two factors interfere with each other. In some cases competition for binding to overlapping sites leads to a mutually exclusive binding. In other cases, factors can bind to DNA simultaneously, but binding of a repressing factor possibly “masks” an activation domain of an activator. A number of molecular mechanisms are suggested for functioning of both synergistic and antagonistic CEs [3].
2. Composite element classification
Classification of the composite elements can be based on different criteria: i) type of interactions between transcription factors involved (synergism or antagonism); ii) structure of transcription factors, namely structure of DNA-binding domains; iii) combinatorial regulation provided by a composite element.
To classify composite elements in terms of factors’ DNA-binding domains we applied a previously suggested transcription factor classification [4]. As shown in the Table 1, the most frequent composite elements include binding sites for bZIP and REL factors and for bZIP and ETS factors.
Table 1. Classification of the composite elements according to the structure of DNA-binding domains of factors interacting (COMPEL release 2.2).
| Class | Factor | |||||||||||||||
| AP-1 | A | 1 | ||||||||||||||
| BZIP | CREB | B | 2 | |||||||||||||
| C/EBP | C | 3 | 1 | |||||||||||||
| BHLH | E2A, MyoD, myogenin | D | 1 | 1 | ||||||||||||
| BHLH-ZIP | TFE3, USF,SREBP, c-Myc, E2F | E | 1 | 3 | ||||||||||||
| NF-1, NF-Y, RF-X | NF-1, NF-Y, RF-X | F | 2 | 1 | 2 | |||||||||||
| C4 zf | GR, ER, PR, RAR, T3R, VDR, HNF-4, COUP | G | 4 | 2 | 2 | 1 | 5 | |||||||||
| C2H2 zf | Sp1, YY1, Egr | H | 1 | 3 | 3 | 2 | ||||||||||
| Diverse C4 zf | GATA | I | 1 | 1 | ||||||||||||
| Homeo domain | Cad, HNF-1 | J | 1 | 1 | 1 | 2 | 1 | |||||||||
| Oct (homeo-POU) | K | 2 | 1 | 1 | 2 | 1 | 3 | |||||||||
| Winged helix | HNF-3 | L | 1 | 3 | 1 | 1 | ||||||||||
| Tryptophan clusters | ETS | M | 11 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | ||||||
| Myb, IRF | N | 2 | 3 | |||||||||||||
| REL homology region | NF-k B, NF-AT | O | 16 | 8 | 2 | 2 | 1 | |||||||||
| MADS box | SRF, MEF2 | P | 1 | 2 | 3 | |||||||||||
| HMG I(Y) | HMG I(Y) | R | 3 | 1 | 1 | 5 | ||||||||||
| RUNT | PEBP2 | S | 2 | 3 | 1 | |||||||||||
|
Code |
A | B | C | D | E | F | G | H | J | K | L | M | N | O | ||
Since functional properties and tissue distribution of factors vary significantly within the same factor class, another criteria for classification is suggested based on combinatorial regulation provided by a CE [3]. Combinatorial regulation may be exemplified by a CE that contains binding sites for c-Ets-1 and Pit-1 (COMPEL acc C00131). c-Ets-1 is ubiquitously distributed factor inducible through Ras/Raf signal transduction pathway, and Pit-1 is expressed exclusively in the pituitary gland. Cooperation of these two factors on the composite element results in synergistic transcriptional activation of prolactin gene within pituitary cells in response to Ras [5].
Based on the combinatorial regulation provided, CEs can be divided into following groups [3]: a) CEs providing tissue-specific regulation, when one factor is tissue- or cell type-specific, and another is ubiquitous and constitutive; b) tissue-specific induction, when one factor is tissue-specific and another is ubiquitous and inducible; c) inducible regulation, when one factor mediates a responce to an extracellular signal and another is ubiquitous and constitutive; d) cross-coupling of signal transduction pathways, in case when both factors are inducible through different pathways; e) cell cycle-dependent regulation, when activity of the first factor is dependent on the cell cycle stage, and second factor is cell-cycle independent.
3. Model of relational database
To describe in a database a complex biological object such as a composite element we should keep information about sites on DNA and factors binding to them as well as experimental evidence confirming synergistic or antagonistic regulation.
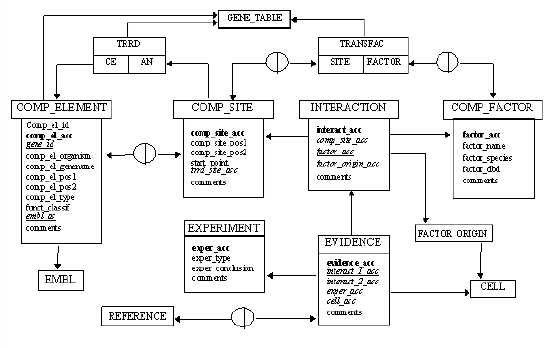
Our relational model for the COMPEL database comprises six main tables in terms of biological subject: comp_element, comp_site, comp_factor, interaction, evidence, and experiment.
The table of comp_elements contains its identifier comp_el_acc and accession number comp_el_acc, description of a gene where a CE is identified (comp_el_organism and comp_el_genename), the first and the last positions of a CE in gene (comp_el_pos1 and comp_el_pos2), information about synergistic or antagonistic type (comp_el_type), and functional classification of a composite element (comp_el_classif). Of importance, this table is linked to EMBL via field embl_ac, and through a field gene_id to the Gene_Table which is common for TRANSFAC and TRRD databases [6-8]. The table comp_elements is connected with comp_site by a n:m relation. By definition, each CE contains two sites, and in some cases the same site may be part of different composite elements.
The table of individual sites making up composite elements, comp_site, contains comp_site_acc, and location of a site in a gene (comp_site_pos1 and comp_site_pos2). Field start_point indicates the reference point for position numbering. In most cases this point is the transcription start site, but in several cases the beginning of an enhancer, or just beginning of a sequence are used.
The field trrd_site_acc links to the TRRD database, and a link to the TRANSFAC SITE Table is provided by a n:m relation.
The table comp_factor contains the fields factor_acc, factor_name, factor_species. The field factor_dbd presents information about the type of DNA-binding domain. This table is connected with TRANSFAC FACTOR Table by a n:m relation, due to dimeric organization of some factors. For example, factor human c-Jun/c-Fos is to be connected with both human c-Jun and human c-Fos in TRANSFAC. From the other hand, human c-Jun may be part of various heterodimeric factors such as c-Jun/c-Fos, c-Jun/FosB, c-Jun/ATF-2 and therefore should be connected with several factors in COMPEL.
The table interaction serves to connect a site with an interacting factor. Each record in this table contains its accession number interact_acc, accession numbers for the corresponding site and factor (comp_site_acc and comp_factor_acc) as well as the field factor_origin_acc for connection with the factor_origin table (endogenous, recombinant, purified, fusion, or in vitro translated factor). Therefor the interaction table allows to connect a site with a certain factor several times depending on the origin of this factor. Each of such interactions has its own accession number by which it is connected with a particular experiment and reference.
Figure 2. A block-diagram representing relational model of the COMPEL database. Links to TRANSFAC and TRRD are shown schematically to SITE and FACTOR tables in TRANSFAC and to CE and AN fields in TRRD. Circles represent linking tables used for establishing n:m relations between the main tables.
The table evidence contains information about experimental evidences for the composite elements. One record in this table contains experimental evidences for one particular pair of factors binding to a pair of sites. Each record has its accession number evidence_acc, accession numbers for two corresponding interactions interact_1_acc and interact_2_acc, accession numbers for an experiment and of a cell type which was used for this experiment (exper_acc and cell_acc). This table is connected by a n:m relation with the table references.
The table experiment contains accession numbers of experiments exper_acc, information about type of experiment in the field exper_type, and what is very important, the field exper_conclusion presents information about the conclusion that has been drawn from the experiment. For example, two principally different conclusions about functional synergism or functional antagonism between factors can be drawn from the same type of experiment – co-transfection assay – depending on the concrete design of the experiment and its result.
The structure described allows to store accurately the relevant experimental data: one can know exactly for each composite element not only the names of interacting factors and types of experimental studies, but also with which factor (including factor origin) the concrete experiment was done.
4. Access to the COMPEL database
The COMPEL database is accessible via SRS system (http://transfac.gbf.de/srs5/) or by anonymous ftp (ftp://transfac.gbf.de/pub/databases/compel/ or ftp://bionet.nsc.ru/pub/biology/compel/ ).
Acknowledgments
Different parts of this work were funded by the German Bundesministerium fur Bildung, Wissenschaft, Forschung und Technologie (project no. X224.6), by the Russian Ministry of Sciences and the Siberian Branch of Russian Academy of Sciences, by the North Atlantic Treaty Organisation (grant no. 951149) as well as by BIOBASE Ltd. (grant N98.1).
References
- M.I. Diamond, J.N. Miner, S.K.Yoshinaga and K.R. Yamamoto “Transcription factor interactions: selectors of positive or negative regulation from a single DNA element” Science 249, 1266-1272 (1990)
- O.V. Kel, A.G. Romaschenko, A.E. Kel, E. Wingender and N.A. Kolchanov “A compilation of composite regulatory elements affecting gene transcription in vertebrates” Nucleic Acids Res. 23, 4097-4103 (1995)
- O.V. Kel, A.E. Kel, A.G. Romaschenko, E. Wingender and N.A. Kolchanov “Composite regulatory elements: classification and description in the COMPEL database” Mol. Biol. 31, 498-512 (1997)
- E. Wingender “Classification scheme of eukaryotic transcription factors”, Mol. Biol. 31, 483-497 (1997)
- A.P. Bradford, K.E. Conrad, C. Wasylyk, B. Wasylyk and A. Gutierrez-Hartmann “Functional interaction of c-Ets-1 and GHF-1/Pit-1 mediates Ras activation of pituitary-specific gene expression: mapping of the essential c-Ets-1 domain” Mol. Cell. Biol. 15, 2849-2857(1995)
- A.E. Kel, N.A. Kolchanov, O.V. Kel, A.G. Romashencko, E.A. Ananko, E.V. Ignatieva, T.I. Merkulava, O.A. Podkolodnaya, I.L. Stepanenko, A.V. Kochetov, F.A. Kolpakov, N.L. Podkolodnyi and A.N. Naumochkin “TRRD: Database on Transcription Regulatory Regions of Eukaryotic Genes” Mol Biol 31, 626-636 (1997)
- E. Wingender, A.E. Kel, O.V. Kel, H. Karas, T. Heinemeyer, P. Dietze, R. Knuppel, A.G. Romaschenko and N.A. Kolchanov “TRANSFAC, TRRD and COMPEL: Towards a federated database system on transcriptional regulation” Nucleic Acids Res. 25, 265-268 (1997)
- T. Heinemeyer, E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E.V. Ignatieva, E.A. Ananko, O.A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny and N. A. Kolchanov “Databases on transcription regulation: TRANSFAC,TRRD and COMPEL” Nucleic Acids Res. 26, 362-367, (1998)