FREIER A., HODING M., HOFESTADT R.+, LANGE M, SCHOLZ U.
Otto-von-Guericke-University Magdeburg
Institute for Technical and Business Information Systems
Bioinformatics and Medical Informatics
Universitatsplatz 2, D-39106 Magdeburg, Germany
e-mail: {freier,hoeding,hofestae,mlange,uscholz}@iti.cs.uni-magdeburg.de
+Corresponding author
Keywords: molecular information system, bioinformatics, information fusion, metabolic network control
1. Introduction
Methods of Biotechnology produce a hugh amount of data which must be stored and analyzed. For the analysis different database systems and software-tools are available [1]. For the progress of biotechnology the analysis and understanding of the genotype/phenotype relation is essential. Based on the molecular database systems the analysis of the genotype/phenotype behavior can be supported using information systems, which combine the molecular information fusion and the simulation of metabolic networks. The idea of the fusion of molecular data is not new one. The Ecocyc system of P. Karp [2] was one of the first integrative molecular database systems. Today software tools are available which allow the implementation of powerful integrative analysis tools. Moreover, for the analysis of metabolic networks models and simulation environments are available [3,4]. In our contribution we will present the architecture of the “Magdeburger Molecular Information System” (MMIS), which allows the access and analysis of metabolic networks. One important application of our MMIS is the detection of inborn errors [5].
2. Molecular Information Systems
The molecular data must be stored and analyzed. Database systems for genes and proteins (EMBL, JDDB, GENBANK, PIR) are offering access via internet. Therefore, in the workfield of molecular biology this point of view allows the analysis of metabolic processes. To understand the molecular logic of cells we must be able to analyze metabolic processes in: qualitative and quantitative terms. In this case modeling and simulation [3] are important methods and will influence the domain of medicine and (human) genetics – the microscopic level. All biosynthetic processes are still collected by the Boehringer company [6]. First electronical information systems are available which represent all collected biochemical pathways [6,7]. The main question is: How can we get access to this molecular data and how can we simulate these processes (metabolic pathways)?
Today integrative molecular information systems are available which represent different molecular knowledge (data). The state of the art is shown by P. Karps system Ecocyc [2], which represents the metabolic pathways of E. coli. For every gene or protein within a specific metabolic pathway EcoCyc presents the access to all corresponding genes and/or proteins. Moreover, the electronical information system KEGG [7] represents all biochemical networks and allows the access to the protein and gene database systems via metabolic pathways. However, both systems are based on the idea of the statical representation of the molecular data and knowledge. The next important step is to implement and integrate powerful interactive simulation environments which allow the access to different molecular database systems and the simulation of complex biochemical reactions. These information systems can support the progress of biotechnology as well as the molecular diagnostic processes.
3. Integration
The integration of heterogeneous data sources is one important aspect of the project. First analysis of available systems show a wide variety of used modeling concepts. However, many relations between the system can be found by the designer of the integrated system. On one hand, we detect some useful standardized global identifiers, e.g. the MIM number for diseases or the EC number for enzymes. On the other hand, we find data, which have a well-known semantics, defined by databases which are already integrated.
Hence our system is partially based on the design of federated databases systems (FDBS), it is appropriate to use available tools for FDBS design, e.g. the SIGMA-Bench [8]. Moreover, the results of the research on semi-structured data can support the integration of semi-structured and WWW-based data source. First experiments show the usability of the FLORID-tool [9]. In that way, the integration of an adequate number of different biomolecular databases is possible with a reasonable amount of work.
4. Magdeburger Molecular Information System
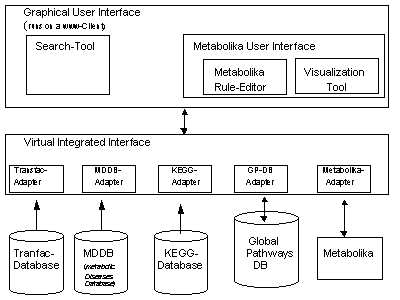
The development of the Magdeburger Molecular Information System (MMIS) is the goal of our project. The architecture of our prototype allows the access onto two different molecular database systems which allow the analysis of metabolic pathways. The access to the molecular knowledge (genes, proteins, and pathways) is realized by using the information system KEGG [7] which allows the access to every known metabolic pathway including the related genes and proteins. Information about the gene regulation is available via the TRANSFAC [10] database system. Our WWW-Server connects both molecular database systems. This integration tool represents the kernel of our MMIS. Furthermore, our system offers the simulation tool Metabolika for the analysis of metabolic pathways. Metabolika allows the interactive simulation of biochemical networks [11]. Therefore, molecular knowledge can be transfered into analytical metabolic rules – the language of Metabolika. Based on that information transfer, the simulation of complex metabolic networks is available. The configuration of Metabolika is represented by the actual metabolite concentrations of the virtual biochemical reaction space. Metabolika allows the calculation of (all) possible configurations (derivation tree) based on the selected metabolic knowledge (biochemical scenario) and the start configuration. The visualization tool and the Graphical User Interface (GUI) realize the interactive analysis of the corresponding derivation tree.
The idea of our MMIS [5] is to present a virtual laboratory for the analysis of molecular processes. Therefore, we integrated different database systems which represent molecular and medical knowledge. The graphical user interface gives the user access to a compact local information system. The access to the molecular knowledge will be realized by the direct access to the heterogeneous database systems. In case of modeling and simulation of metabolic processes the specific biochemical knowledge will be identified by using these database systems. In the next step this knowledge will be transfered automatically into the language of analytical metabolic rules, the language of Metabolika. The simulation of this biochemical reactions will be produced by Metabolika. For the visualization and statistical analysis of the derivation tree tools are available.
5. Application
Metabolic diseases are caused by genetic defects [12]. New methods of biotechnology support the diagnosis (molecular diagnosis) of metabolic diseases, and gene therapy is becoming popular. Regarding the detection of inborn errors, the first database system METAGENE [13] which represents macroscopic knowledge (medical knowledge) is available. The idea of another project in our workgroup is to expand the macroscopic knowledge by collecting molecular information (microscopic knowledge). Therefore, we have developed and implemented the Metabolic Disease DataBase (MDDB) [14]. The key idea of this information system is to combine microscopic and macroscopic knowledge of inborn errors.
However, we already implemented a Molecular Information System for the analysis of inborn errors. Therefore we expand the architecture of our system by the integration of the MDDB database system.
Regarding metabolic diseases, our molecular information system presents information about the corresponding metabolic pathways, genes, enzymes, and medical knowledge. Moreover, relevant references and therapy plans will be offered.
Figure 1: The architecture of MMIS
6. Conclusion
Different molecular database systems are available which represent genes, proteins, and metabolic pathways [2]. The state of the art in the field of molecular bioinformatics is moving to the implementation of integrative database systems [2,7] in combination with dynamic simulation environments for the analysis and synthesis of metabolic networks [5].
The detection of metabolic diseases is one important application of molecular information systems, which represent molecular and medical data and allow the simulation of metabolic processes. Nearly one of every 800 newborns carries an inborn error [12]. Most of these defects cannot be identified. The early detection of inborn errors is vital for the therapy of these diseases. Therefore, database systems representing this medical information are necessary. We have already implemented an integrative molecular information system for the detection of inborn errors based on microscopic (molecular) and macroscopic (medicine) knowledge. This system allows the integrative access of medical and molecular knowledge connecting different database systems. Based on our integrative meta-database system, macrosopic and microscopic knowledge can be selected. Our rule based system Metabolika allows the interactive microscopic analysis of biochemical networks [11,15].
Acknowledgement
This work is supported by the Kurt-Eberhard-Bode-Stiftung (grant Bioinformatik and Medizinische Informatik) im Stifterverband fur die Deutsche Wissenschaft.
References
- S. Suhai (ed.), “Theoretical and Computational Methods in genome Research” (Plenum Press, New York, 1997)
- P. Karp, “A knowledge base of the chemical compounds of intermediary metabolism” CABIOS 8 (4), 347-357 (1992)
- R. Hofestadt, J. Collado-Vides, M. Mavrovouniotis, “Modeling and Simulation of Metabolic Pathways, Gene Regulation and Cell Differentiation” BioEssays, 18 (4), 333-335 (1996)
- R. Hofestadt, “Theorie der regelbasierten Modellierung des Zellstoffwechsels” (Shaker Verlag, Aachen, 1996)
- R. Hofestadt, U. Scholz, “Information processing for the analysis of metabolic pathways and inborn errors” BioSystems, in press (1998)
- G. Michael, “Biochemical Pathways” (Boehringer, Mannheim, 1993)
- S. Goto, H. Bono, H. Ogata, T. Fujibuchi, T. Nishioka, K. Sato, M. Kanehisa, “Organizing and computing metabolic pathway data in terms of binary relations” In R. Altman, A. Dunker, L. Hunter (eds.), “Proceedings of the Pacific Symposium on Biocomputing” pp. 175-186 (World Scientific, Singapore,1997)
- S. Conrad, M. Hoding, G. Saake, I. Schmitt, C. Turker, “Schema Integration with Integrity Constraints” In C. Small, P. Douglas, R. Johnson, P. King, N. Martin, (eds.), “15th British National Conf. on Databases, BNCOD 15, London, UK”, pp. 200-214, LNCS 1271, (Springer-Verlag, Berlin, 1997)
- R. Himmeroder, G. Lausen, B. Ludascher, C. Schlepphorst, “FLORID: A DOOD-System for Querying the Web” Demonstration Session at EDBT’98, (Valencia, Spain, 1998)
- E. Wingender, P. Dietze, H. Karas, R. Knuppel, “TRANSFAC: A database on transcription factors and their DNA binding sites” Nucleic Acids Research 24 (1), 238-241 (1996)
- R. Hofestadt, F. Meinecke, “Interactive Modelling and Simulation of Biochemical Networks” Computers in Biology and Medicine 25, 321-324 (1995)
- R. Trent, “Molekulare Medizin” (Spektrum Akademischer Verlag, 1993)
- U. Mischke, G. Frauendienst-Egger, P. Matthis, P. Gao, F.-K. Trefz, “KBS-DIAMET: database and expert system for diagnosis and treatment of patients with inborn errors of metabolism” J. Inher. Metab. Dis. 18, 224-226 (1995)
- R. Hofestadt, M. Pru?, U. Scholz, H. Urban, “The Metabolic Diseases DataBase: The First Molecular Information System for the Detection of Inborn Errors”, submitted
- R. Hofestadt, “A Rule Based System for the Detection of Metabolic Diseases” In R. Green, H. Peterson, D. Protti (eds.), “Proceedings of the 8th World Congress Medical Informatics” pp. 994-999 (North-Holland,1995)