TOMITA MASARU1, KENTA HASHIMOTO1, KOUICHI TAKAHASHI1, TOM SHIMIZU1, YURI MATSUZAKI1, FUMIHIKO MIYOSHI1, KANAKO SAITO1, SAKURA TANIDA1, KATSUYUKI YUGI1, J. CRAIG VENTER2, CLYDE A. HUTCHISON2
1Laboratory for Bioinformatics, Keio University
2The Institute for Genomic Research
Keywords: procariote model cell, virtual whole cell simulation, mycoplasma genitalium
A procaryote model cell has been constructed using E-CELL, a computer software environment developed for conducting virtual whole cell simulation. The genome of this hypothetical cell currently consists of 127 genes including 20 tRNA genes and 2 rRNA genes. The gene set was derived by combining pathways from Mycoplasma genitalium, the self replicating organism with the smallest known gene set. These pathways constitute the fundamental basis of cellular metabolism, driven by ATP synthesized via glycolysis. The virtual cell produces membrane phospholipids and expresses genes through transcription and translation.
We developed E-CELL, a generic computer software environment for modeling and simulation of whole cell systems. E-CELL is an object-oriented environment for simulating molecular processes in user-definable models, equipped with interfaces that allow observation and intervention. Using E-CELL, we constructed a model of a hypothetical cell with only 127 genes sufficient for transcription, translation, energy production and phospholipid synthesis.
The E-CELL system is implemented as a object oriented simulation system written in C++. The simulation engine, cell model, and interfaces of the E-CELL system are realized as independent software objects, allowing flexible development. The cell model is defined as a list of two fundamental entities: substances and reaction rules. Typical substances include proteins, protein complexes, RNA, and small molecules such as glucose and amino acids. Reaction rules define the reactions which can take place within the cell. The state of the cell at each time interval is expressed as a list of quantities of all substances within the cell, along with global values such as cell volume, pH and temperature. The quantity of a substance is defined as the number of molecules, and is represented internally by an integer. Quantity can be easily converted to concentration by referring to the volume.
The simulator engine generates the next state in time by pseudo-parallel computation of all functions defined in the reaction rules. Each rule is called upon by the simulator engine to compute the quantity of each substance in the next time unit. The net change in quantity for each substance is calculated, generating the next state of the cell.
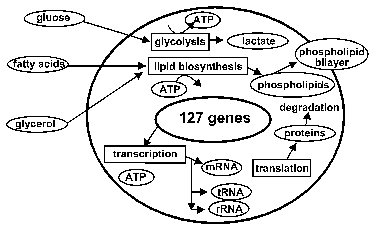
The hypothetical cell we have modeled uptakes glucose from the culture medium using a phosphotransferase system, generates ATPs by catabolizing glucose to lactate by glycolysis and fermentation pathways, and exports lactate out of the cell. Since enzymes and other proteins are modeled to degrade spontaneously over time, they must be constantly synthesized in order for the cell to sustain “life”. The protein synthesis is implemented by modeling the molecules necessary for transcription and translation, namely RNA polymerase, ribosomal subunits, rRNAs, tRNAs and tRNA ligases. The cell also uptakes glycerol and fatty acid and produces phosphatidyl glycerol for membrane structure using a phospholipid biosynthesis pathway (figure 1).
| Gene type | M.gen | Other | Total |
| Glycolysis | 9 | 0 | 9 |
| Lactate fermentation | 1 | 0 | 1 |
| Phospholipid biosynthesis | 4 | 4 | 8 |
| Phosophotransferase system | 2 | 0 | 2 |
| Glycerol uptake | 1 | 0 | 1 |
| RNA polymerase | 6 | 2 | 8 |
| Amino acid metabolism | 2 | 0 | 2 |
| Ribosomal L subunit | 30 | 0 | 30 |
| Ribosomal S subunit | 19 | 0 | 19 |
| rRNA | 2 | 0 | 2 |
| tRNA | 20 | 0 | 20 |
| tRNA ligase | 19 | 1 | 20 |
| Initiation factor | 4 | 0 | 4 |
| Elongation factor | 1 | 0 | 1 |
| Protein coding genes | 98 | 7 | 105 |
| RNA coding genes | 22 | 0 | 22 |
| Total | 120 | 7 | 127 |
Table 1: The number of genes important pathways of the hypothetical cell. Most of the genes are taken from M. Genitalium and are listed in the column “M.gen”. Genes not found in M. genitalium which were taken from other organisms such as E.coli are listed in the column “Other”.
The E-CELL system provides a number of graphical interfaces which allow the user to observe the cell’s state and manipulate it interactively). The E-CELL interfaces provide a means of conducting “experiments in silico”. For example, we can “starve” the cell by draining glucose from the culture medium. The cell would eventually “die” running out of ATP. If glucose is added back, it may or may not recover depending on the starvation duration. We can also “kill” the cell by knocking out an essential gene for, e.g., protein synthesis. The cell would become unable to synthesize proteins, and all enzymes would eventually disappear due to spontaneous degradation.
In conclusion, our simulation work with E-CELL has shown that modeling cellular metabolisms as a whole cell appears feasible by defining substances and reaction rules. Whether or not complete living cells could be modeled and simulated at the molecular level is still an open question. However, the rapidly increasing information in genomics and molecular biology increase the likelihood that whole cell simulation of real organisms will be a feasible task in the near future.
Acknowledgements
This work was supported by Eisai Research Institute and also in part by a Grant-in-Aid for Scientific Research on Priority Areas ‘Genome Science’ from The Ministry of Education and Science in Japan.
| ID | name | ID | name |
| EC1.1.1.27 | L-Lactate dehydrogenase | EC6.1.1.16 | Cysteine–tRNA ligase |
| EC1.2.1.12 | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | EC6.1.1.17 | Glutamate–tRNA ligase |
| EC2.1.2.9 | Methionyl-tRNA formyltransferase | EC6.1.1.18 | Glutamine–tRNA ligase |
| EC2.7.1.107 | Diacylglycerol kinase | EC6.1.1.19 | Arginine–tRNA ligase |
| EC2.7.1.11 | 6-Phosphofructasokinase | EC6.1.1.2 | Tryptophan–tRNA ligase |
| EC2.7.1.30 | Glycerol kinase | EC6.1.1.20 | Phenylalanine–tRNA ligase |
| EC2.7.1.40 | Pyruvate kinase | EC6.1.1.21 | Histidine–tRNA ligase |
| EC2.7.1.69 | phosphotransferasesystem enzyme II, ABC component(ptsG) | EC6.1.1.22 | Asparagine–tRNA ligase |
| EC2.7.2.3 | Phosphoglycerate kinase | EC6.1.1.3 | Threonine–tRNA ligase |
| EC2.7.3.9 | phosphoenolpyruvate-proteinphosphotransferase(ptsI) | EC6.1.1.4 | Leucine–tRNA ligase |
| EC2.7.4.4 | Nucleoside-phosphate kinase | EC6.1.1.5 | Isoleucine–tRNA ligase |
| EC2.7.4.6 | Nucleoside-diphosphate kinase | EC6.1.1.6 | Lysine–tRNA ligase |
| EC2.7.7.41 | CDPdiglyceride pyrophosphorylase | EC6.1.1.7 | Alanine–tRNA ligase |
| EC2.7.8.5 | CDPdiacylglycerol-glycerol-3-phsophate 3-phosphatidyltransferase | EC6.1.1.9 | Valine–tRNA ligase |
| EC3.1.1.23 | Acylglycerol lipase | EC5.3.1.9 | Glucose-6-phosphate isomerase |
| EC3.1.1.3 | Lipase | EC5.4.2.1 | Phosphoglycerate mutase |
| EC3.1.3.21 | Glycerol-1-phosphatase | EC6.1.1.1 | Tyrosine–tRNA ligase |
| EC3.1.3.27 | Phosphatidylglycerophosphatase | EC6.1.1.10 | Methionine–tRNA ligase |
| EC3.6.1.1 | Inorganic pyrophosphatase | EC6.1.1.11 | Serine–tRNA ligase |
| EC3.6.1.1 | Pyrophosphatase | EC6.1.1.12 | Aspartate–tRNA ligase |
| EC4.1.2.13 | Fructose-bisphosphate aldolase | EC6.1.1.14 | Glycine–tRNA ligase |
| EC4.2.1.11 | Phosphopyruvate hydratase | EC6.1.1.15 | Proline–tRNA ligase |
| EC5.3.1.1 | Triose-phosphate isomerase |
Table 2: Enzymes in the hypothetical cell
| MG005 | Serine–tRNA ligase | MG153 | ribosomal protein L23 |
| ID | name | ID | name |
| MG021 | Methionine–tRNA ligase | MG215 | 6-phosphofructokinase (pfkA) |
| MG023 | fructose-bisphosphate aldolase (tsr) | MG216 | pyruvate kinase (pyk) |
| MG033 | glycerol uptake facilitator(glpF) | MG232 | ribosomal protein L21 |
| MG035 | Histidine–tRNA ligase | MG234 | ribosomal protein L27 |
| MG036 | Aspartate–tRNA ligase | MG249 | RNA polymerase sigma S subunit |
| MG038 | glycerol kinase (glpK) | MG251 | Glycine–tRNA ligase |
| MG041 | Protein histidine(HPr)(ptsH) | MG253 | Cysteine–tRNA ligase |
| MG069 | phosphotransferase enzymeII(ptsG) | MG257 | ribosomal protein L31 |
| MG070 | ribosomal protein S2 | MG266 | Leucine–tRNA ligase |
| MG081 | ribosomal protein L11 | MG283 | Proline–tRNA ligase |
| MG082 | ribosomal protein L1 | MG292 | Alanine–tRNA ligase |
| MG087 | ribosomal protein S12 | MG300 | phosphoglycerate kinase (pgk) |
| MG088 | ribosomal protein S7 | MG301 | G3PD (gapA) |
| MG089 | Elongation Factor G | MG311 | ribosomal protein S4 |
| MG090 | ribosomal protein S6 | MG325 | ribosomal protein L33 |
| MG092 | ribosomal protein S18 | MG334 | Valine–tRNA ligase |
| MG093 | ribosomal protein L9 | MG340 | RNA polymerase beta’ subunit |
| MG111 | phosphoglucose isomerase B (pgiB) | MG341 | RNA polymerase beta subunit |
| MG113 | Asparagine–tRNA ligase | MG344 | Lipase |
| MG114 | PGP synthase (pgsA) | MG345 | Isoleucine–tRNA ligase |
| MG126 | Tryptophan–tRNA ligase | MG351 | inorganic pyrophosphate (ppa) |
| MG136 | Lysine–tRNA ligase | MG361 | ribosomal protein L10 |
| MG142 | translation initiation factor2 | MG362 | ribosomal protein L7 |
| MG150 | ribosomal protein S10 | MG363 | ribosomal protein L32 |
| MG151 | ribosomal protein L3 | MG363.1 | ribosomal protein S20 |
| MG152 | ribosomal protein L4 | MG365 | Methionyl-tRNA formyltransferase |
| MG154 | ribosomal protein L2 | MG375 | Threonine–tRNA ligase |
| MG155 | ribosomal protein S19 | MG378 | Arginine–tRNA ligase |
| MG156 | ribosomal protein L22 | MG407 | enolase (eno) |
| MG157 | ribosomal protein S3 | MG417 | ribosomal protein S9 |
| MG158 | ribosomal protein L16 | MG418 | ribosomal protein L13 |
| MG159 | ribosomal protein L29 | MG424 | ribosomal protein S15 |
| MG160 | ribosomal protein S17 | MG426 | ribosomal protein L28 |
| MG161 | ribosomal protein L14 | MG429 | proteinphosphotransferase(ptsI) |
| MG162 | ribosomal protein L24 | MG430 | phosphoglycerate mutase (pgm) |
| MG163 | ribosomal protein L5 | MG431 | triosephosphate isomerase (tpiA) |
| MG164 | ribosomal protein S14 | MG433 | Transcription elongation factor Ts |
| MG165 | ribosomal protein S8 | MG437 | CDP-diglyceride synthetase (cdsA) |
| MG166 | ribosomal protein L6 | MG444 | ribosomal protein L19 |
| MG167 | ribosomal protein L18 | MG446 | ribosomal protein S16 |
| MG168 | ribosomal protein S5 | MG451 | Transcription elongation factor Tu |
| MG173 | translation initiation factor1 | MG455 | Tyrosine–tRNA ligase |
| MG174 | ribosomal protein L36 | MG460 | L-lactate dehydrogenase (ldh) |
| MG175 | ribosomal protein S13 | MG462 | Glutamate–tRNA ligase |
| MG176 | ribosomal protein S11 | MG466 | ribosomal protein L34 |
| MG177 | RNA polymerase alpha core subunit | SCMNPK | Nucleoside-phosphate kinase |
| MG178 | ribosomal protein L17 | ECNDK | Nucleoside-diphosphate kinase |
| MG194 | Phenylalanine–tRNA ligase alpha | ECGLNS | Glutamine–tRNA ligase |
| MG196 | transltion initiation factor3 | T0001 | Acylglycerol lipase |
| MG197 | ribosomal protein L35 | T0002 | Glycerol-1-phosphatase |
| MG198 | ribosomal protein L20 | ECPGPB | Phosphatidylglycerophosphatase |
| ECDGKA | Diacylglycerol kinase (dgkA) |
Table 3: Protein coding genes in the hypothetical cell. 120 of the 127 genes, given ID’s beginning with “MG”, are present in the genome of M. genitalium. ID’s starting with “EC” represent genes present in E. coli. Genes for nucleoside-phosphate kinase, Acylglycerol lipase, and Glycerol-1-phosphatase are not found in either M. genitalium or E. coli. The nucleoside-phosphate gene was given the ID “SCMNPK” because it has been sequenced in Schistosoma mansoni. ID’s starting with “T”(temporary) were assigned for acylglycerol lipase and Glycerol-1-phosphatase because no sequences for either gene have been submitted to GenBank.
Figure 1: Metabolism overview of the model cell. It has pathways for glycolysis and phospholipid biosynthesis, as well as transcription and translation metabolisms.