Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 10 Lavrentiev Ave., Novosibirsk, 630090, Russia; e-mail: peter@bionet.nsc.ru;
+Corresponding author
Keywords: eukaryotic promoters, nucleotide frequency profiles, CpG islands, sequence complexity, TATA-box, DNA-bending stiffness
Introduction
Eukaryotic promoters are DNA sequences providing gene expression regulation at the stage of transcription initiation. They have a complex block-modular structure and contain numerous short functional elements, transcription factor binding sites.
The distal elements have no exact uniform location, are dispersed in 5′-flanking region up to ~1 kb upstream of the transcription start site (TSS), and are involved in specific transcription regulation (tissue-, cell-specific, etc.).
The proximal elements, which encompass TSS directly, are called core elements and are involved in formation of the basal transcription complex. Belonging to them are TATA box with consensus sequence TATA(A/T)A(A/T) and Inr with consensus sequence YYAN(T/A)YY. Transcription initiation begins with formation of the basal transcription complex in the promoter region of several dozens bp long located around the TSS. In turn, assembling of the basal transcription complex at TATA-containing promoters starts with the recognition of TATA boxes by TATA-binding protein (TBP) [1].
The goal of this work was to study the peculiarities of the nucleotide context and the DNA conformational properties influencing the TBP/TATA binding.
Materials and methods
Materials
Sequences of nonhomologous promoters of vertebrate chromosome genes were extracted from EPD42 [2] using software package MGL [3]. Each sequence represented a [-300; +100] region relative to the TSS at position +1.
Construction of position-specific nucleotide frequency profiles over the sample
The number of nucleotides of a definite type Nia (i, the number of position; a, type of nucleotide in 15 single letter-based code [4]) was calculated over the sample of sequences in each position from -300 to +100; the relative frequencies Nia/N (N, sample size) were plotted vs. every position. The resulting graphs are the position-specific nucleotide frequency profiles along the region [-300; +100] over the sample of sequences.
Sorting of TATA-containing (TATA+) and TATA-less (TATA-) promoters
The weight matrix for TATA box with a cut-off value of -8.16 [5] was used to separate the initial sample into TATA+ and TATA- subsamples.
Sorting promoters into CpG+ and CpG- subsamples
Cytosine residues in CpG dinucleotides are methylated in the major part of vertebrate DNA. However, there are specific DNA regions, the so-called CpG islands, where the CpG dinucleotides are nonmethylated in all the tissues. Characteristic of these DNA islands is the length of over 200 nucleotides, over 50% G+C content, and the ratio of observed/expected (Obs./Exp.) CpG dinucleotides >0.6.
Here we determined the boundaries of CpG islands as follows. Obs./Exp. ratio was calculated as:
![]() ,
,
where L is the length of the sequence in question. Obs./Exp. CpG and (G+C)% were determined in a window of 100 nucleotides long (L=100) moved along the sequence with one-nucleotide shift. The overlapping windows with (C+G)% over 50 and Obs./Exp. CpG over 0.6 were merged; in case the resulting CG-rich fragment exceeded 200 nucleotides, it was considered CpG island.
The promoters containing CpG islands and starting in the 5′ region relative to TSS composed the CpG+ subsample. In addition, several promoters, the left boundary of which was upstream of the TSS and right boundary located downstream of position +95, were included into the CpG+ subsample due to the insufficient length (100 nucleotides) of 3′ DNA region preventing the revelation of all the promoters containing CpG islands that start in the 5′-upstream region from the TSS. The rest promoters formed the CpG- subsample.
Estimation of nucleotide sequence complexity
Complexity of a nucleotide sequence is defined as the least number of events required to generate this sequence [6]. The events are (1) generation of a new symbol and (2) replication of the already generated symbol in certain orientation: direct (D), symmetrical (S), or inverse (I). A definite set of orientations for replication may be specified, for example, replication in only one of the orientations (D-, S-, or I-complexities) or in any orientations (DSI-complexity) may be permitted.
Results and discussion
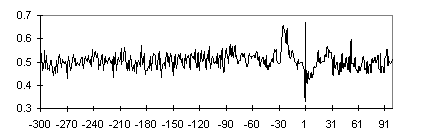
The position-specific nucleotide frequency profiles in 15 single letter-based code were constructed for vertebrate promoters. The regions corresponding to the core elements (TATA box and Inr) differed in their nucleotide context. The W(A+T) nucleotide frequency profile indicates an increased concentration of these nucleotides in the TATA box region (Fig. 1). The transition from pyrimidine (-1) to purine (+1) on the background of a “purine pit” is well evident (Fig. 2).
Fig. 1. The frequency profile of W(A+T) nucleotides for the TATA+ promoter subsample. Position relative to the TSS is plotted on the abscissa; nucleotide frequency in this position over the sample of sequences, on the ordinate.
Fig. 2. The frequency profile of R(A+G) nucleotides for TATA+ promoter subsample. See caption to Fig. 1.
Note the monotonic increase in the concentration of S(G+C) nucleotides with approaching the TATA box region from the 5′ side in the region ~[-300,-100] and higher frequencies of S nucleotides in the regions flanking TATA box. Is this behavior of G,C frequencies typical of individual promoters or an integral property typical of the sample? High expressed genes including housekeeping genes as well as certain genes with limited or tissue-specific expression are known to be associated with CpG islands starting in the 5′ region relative to the TSS. Such islands, nonmethylated DNA regions with a higher content of CpG dinucleotides relative to the remainder of the genome, are typical of vertebrate genes [7]. The boundaries of CpG islands is 5′ region are variable relative to the TSS, and it could have affected the nucleotide frequency profile constructed over all the 301 promoters.
The initial sample of promoters was used to construct subsamples TATA+, TATA-, CpG+, and CpG- and their intersections TATA+CpG+, TATA+CpG-, TATA-CpG+, and TATA-CpG-. CpG+ contains approximately equal number of TATA-containing and TATA-less promoters (70 and 74, respectively), whereas TATA-containing promoters are predominant in CpG- subsample (124 vs. 33 TATA-less; Table 1). Nucleotide composition of the subsamples is listed in Table 2. The locations of CpG islands relative to the TSS in the 5′ region have been determined for promoters from CpG+ subsample; the data are summarized in Fig. 3. The 5′ boundaries of CpG islands relative to the TSS in the 5′ region are distributed evenly. This effect explains the monotonic increase in the C+G frequencies in the corresponding profile for the CpG+TATA+ subsample (Fig. 4). However, the regions with increased G+C content flank the TATA box region also in the G+C frequency profile of CpG-TATA+ (Fig. 5).
Table 1. Number of promoters in the subsamples
|
CpG+ |
CpG- |
||
|
TATA+ |
70 |
124 |
194 |
|
TATA- |
74 |
33 |
107 |
|
144 |
157 |
Table 2. Promoter nucleotide composition in the subsamples
|
CpG+ TATA+ |
CpG+ TATA- |
CpG- TATA+ |
CpG- TATA- |
|
|
A |
0.19 |
0.18 |
0.26 |
0.25 |
|
T |
0.18 |
0.19 |
0.25 |
0.25 |
|
G |
0.31 |
0.32 |
0.24 |
0.25 |
|
C |
0.32 |
0.31 |
0.25 |
0.25 |
Fig. 3. Concentration of CpG islands increases with approaching the TSS. Position relative to the TSS (+1) is plotted on the abscissa; number of promoters containing CpG islands that start in the 5’-upstream region from the TSS, on the ordinate.
Fig. 4. Frequency profile of S(G+C) nucleotides for TATA+CpG+ promoter subsample. See caption to Fig. 1.
Fig. 5. Frequency profile of S(G+C) nucleotides for TATA+CpG- promoter subsample. See caption to Fig. 1.
The C+G-rich regions flanking TATA box may be involved in the binding of TATA-binding protein (TBP) with TATA box. The process of TBP/TATA-binding consists of the following stages [8, 9]: nucleosome displacement, nonspecific TBP binding to DNA, TBP diffusion along the DNA; and site-specific TBP binding to TATA box in the minor groove, causing a 80А DNA bend towards the major groove.
Since the TBP interaction with the DNA minor groove occurs mainly due to the van der Waals interactions, TBP exhibits an increased affinity for AT-rich DNA regions with smooth minor groove along with the decreased affinity for GC-rich regions due to the NH2 group of guanine, which projects into the minor groove and prevents the close contact of protein and DNA surfaces. Thus, the decreased TBP affinity for CG regions does not preclude from its diffusion toward TATA box. In addition, the significant widening of the TATA-box minor groove, wherein it interacts with TBP, was also demonstrated [10].
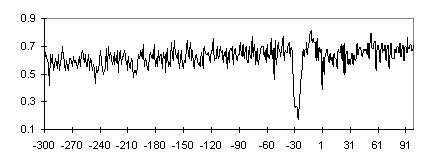
It was demonstrated that TBP bound to the DNA pre-bent (17А-20А ) toward the major groove with a manifold increased affinity compared with the unbent DNA [11]. The promoter DNA architecture may be changed, for example, by activators or HMG-box proteins, which bend DNA, or superhelical coils produced by topoisomerases, etc. It may be suggested that under the stresses causing DNA bending, the stiff CG blocks flanking the TATA box provide the precise location of the bend in AT-rich DNA region located between them and, consequently, the location of TATA box. The bending stiffness profile constructed over the TATA+ subsample is shown in Fig. 6.
Fig. 6. Bending stiffness profile for TATA+ promoter subsample. Position relative to the TSS (+1) is plotted on the abscissa; persistent length, on the ordinate.
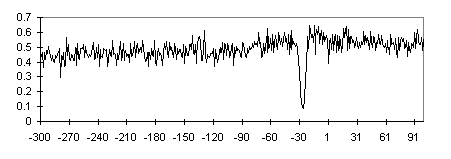
Complexity of nucleotide sequences [6] was estimated for promoter fragments of 20 bp. The TATA-containing regions appeared to be the most complex compared with the other fragments of the same promoters. The S-complexity profile constructed over the TATA+ subsample is shown in Fig. 7. The increased S-complexity of the TATA-containing regions indicates their asymmetry. The asymmetry demonstrated may be related to the correct orientation of TBP on TATA box, when the C-terminus interacts with the first half of the TATA box.
Fig. 7. The S-complexity profile for the TATA+ promoter subsample. Position relative to the TSS (+1) is plotted on the abscissa; total complexity of the 20 bp fragments with the centers in this position calculated over the promoter subsample, on the ordinate.
Analysis of the conformational characteristics has demonstrated that the DNA twist values in the TATA-box region are decreased [10], whereas the increased values are characteristic of the nucleosome positioning sites [10], suggesting a more facile nucleosome displacement near the TATA box.
The interaction of TBP with TATA box is the first, and, probably, most limiting stage in assembling the basal transcription complex at the TATA-containing promoters. Hence the promoters have evolutionary acquired the context peculiarities that allow the optimization of this process at every stage: nucleosome displacement, diffusion of the protein along the DNA, and binding to TATA box. Interaction of TBP with TATA box at a given level may be regulated by alteration of the context.
Acknowledgments
We are grateful to N.A. Kolchanov and F.A. Kolpakov for helpful discussions.
This work was supported by grants from the Russian Foundation for Basic Research (No.97-04-49740, 97-07-90309, 96-04-50006, 98-04-49479, 98-07-90126); Russian Ministry of Science and Technologies; Russian Human Genome Project; Russian Ministry of High Education; Siberian Department of RAS (Programms for support of reseach of young scientists and Programm of Integration projects); National Institutes of Health, U.S.A. (No.5-R01-RR-04026-08)
References
- D.S. Latchman, “Eukaryotic transcription factors” (Academic Press Ltd., London, 1995).
- P. Bucher and E.N. Trifonov, “Compilation and analysis of eukaryotic POL ll promoter sequences” Nucleic Acids Res. 14, 10009 (1986).
- F.A. Kolpakov, V.N. Babenko, “A computer system MGL – the tool for samples construction, visualization and analysis of genomic regulatory sequences” Mol. Biol. (Mosk.) 31, 647 (1997).
- A. Cornish-Bowden, “Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984” Nucleic Acids Res. 13, 3021 (1985).
- P. Bucher, “Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences” J. Mol. Biol. 212, 563 (1990).
- V.D. Gusev, V.A. Kulichkov, and O.M. Chupakhina, “Complexity analysis of genomes. Measures of complexity and classification of the structural regulations revealed” Mol. Biol. (Mosk.) 25, 825 (1991).
- M. Gardiner-Garden and M. Frommer, “CpG islands in vertebrate genomes” J. Mol. Biol. 196, 261 (1987).
- R.A. Coleman and B.F. Pugh, “Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA” J. Biol. Chem. 270, 13850 (1995).
- R.A. Coleman, K.P. Taggart, L.R. Benjamin and B.F. Pugh, “Dimerization of the TATA binding protein” J. Biol. Chem. 270, 13842 (1995).
- M.P. Ponomarenko, Yu.V. Ponomarenko, A.E. Kel’, N.A. Kolchanov, H. Karas, E. Wingender, and H. Sklenar, “Computer analysis of DNA conformational peculiarities of eukaryotic promoter TATA boxes” Mol. Biol. (Mosk.), 31, 733-740 (1997).
- J.D. Parvin, R.J. McCormick, P.A. Sharp, and D.E. Fisher, “Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor” Nature 373, 724 (1995).