FROLOV A.S.#, LAVRYUSHEV S.V., VOROBIEV D.G., GRIGOROVICH D.A.
Laboratory of Theoretical Genetics, Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, 630090 Russia
#Corresponding author e-mail: fas@bionet.nsc.ru
Keywords: promoter recognition, database activation, transcription regulation, Internet-based recogniton
The data on regulation of eukaryotic gene expression is being rapidly accumulated. Such well-known databases as EPD [Peter, 1998], TRANSFAC [Wingender, 1996], TRRD [Kel A., 1997], COMPEL [Kel O., 1995b], and EpoDB [Salas, 1998] have been developed.
A great number of WWW servers containing similar databases and programs for molecular genetic studies are available worldwide. However, these servers suggest a list of resources that can be used only independently of one another, whereas a number of molecular genetic problems demand simultaneous employment of several databases and sequential or simultaneous running of several programs. For example, genome annotation requires the search for homology in GenBank and EMBL databases and/or recognition of functional sites by their patterns using databases on transcription regulation. It is evident that a simple hypertext-based integration would not help to solve this problem.
The positive experience of information integration accumulated during development and use of the Sequence Retrieval System (SRS) [Etzold, 1993], the system for operation with molecular biological databases; the data are accessible via the Internet.
Such databases as GenBank, EMBL, TRASFAC, and TRRD are now accessible under SRS. It allows these resources to be used as initial data for creation of the programs for recognition, homology search, etc.
We proposed the system GeneExpress [Kolchanov et al., 1998] as a first step in realization of the integrated approach to analysis of nucleotide sequences.
The system GeneExpress has been designed to integrate description, annotation and recognition of eukaryotic regulatory sequences. The system contains the following basic units: (1) GeneNet contains an object-oriented database for accumulation of data on gene networks and signal transduction pathways and a Java-based viewer that allows an exploration and visualization of the information on gene networks; (2) Transcription Regulation combines the database on transcription regulatory regions of eukaryotic genes (TRRD) and TRRD Viewer; (3) Transcription Factor Binding Site Recognition contains a compilation of transcription factor binding sites (TFBSC) and programs for their analysis and recognition; (4) mRNA Translation is designed for analysis of structural and contextual properties of mRNA 5íUTRs and prediction of their translation efficiency; and (5) ACTIVITY is the module for analysis and site activity prediction of a given nucleotide sequence. Integration of these databases in GeneExpress is based on the Sequence Retrieval System (SRS) created in the European Bioinformatics Institute.
The next step in this direction is a superstructure based on GeneExpress that provides the search for promoters of a given type employing the data accumulated in the databases of TRRD [Kelí et al, 1997] and the recognition methods created basing on sequence sets from the SAMPLES database.
The basic idea on the approach proposed is to used the information compiled in TRRD as ready-to-use scenarios for promoter recognition. In the simplest case, it allows the regions similar to a given promoter to be detected in an arbitrary sequence.
Let’s consider the system operation using this simplest situation.
Let’s consider a promoter P{pi}, for which the following information is contained in TRRD: it contains N known transcription factor binding sites {(an,bn)} with the site boundaries an and bn relative to the transcription start (here pO{ATGC}, 1<=n<=N).
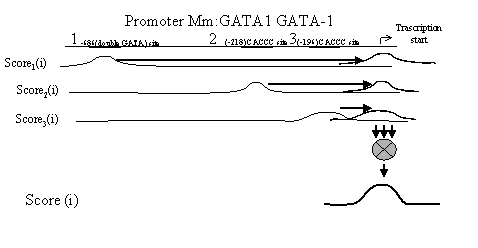
Fig. 1. Illustration of Scoren(i) and Score(i) calculation.
The nucleotide sequence S={si} with the length L (here sŒ{ATGC}, 1<=i<=L) is analyzed to construct the set of N similarity profiles {Scoren(i)} for each nth binding site of this promoter:
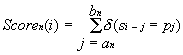 |
(1) |
Equation 1 ascribes to the ith position of the sequence S the number {Scoren(i)} of the coincidences of its region with the boundaries (i-an, ai-bn) with the region of the promoter considered with the boundaries (an,bn), that is, with the binding site of each nth transcription factor. Then the integral similarity profile {Score(i)} of this sequence and entire promoter is constructed:
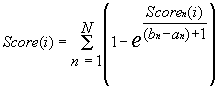 |
(2) |
Equation 2 ascribes to the ith position of the sequence S the value of the similarity to the transcription start of the promoter P considered: the greater is the integral similarity of each of the considered nth binding sites of this promoter to this region of this sequence, the greater is the ascribed value.
Note that in the course of calculations, Scoren(i) is “shifted” along the sequence by a length (an+bn)/2 relative to the nth binding site, so that its maximal value coincides with the transcription start (bold horizontal arrows in Fig. 1). Thus, it is not necessary to consider the positions of concrete binding sites while calculating Scoren(i).
The Scoren(i) is used to predict the potential transcription starts in the sequence S as follows. The mean value M and standard deviation s are calculated and used to find the region with the borders {c,d} within which the value Scoren(i) exceeds the threshold value M+3*s, corresponding to the confidence interval a~0.01 of the Student’s test with the number of degrees of freedom >>100. This region houses the maximal value Score(t), and the position t is predicted as a potential transcription start T of the sequences S. When K such regions {ck,dk} are found, K potential transcription starts {tk} are predicted (here 1<=k<=K).
The system is available at http://wwwmgs.bionet.nsc.ru/Programs/SeqAnn/
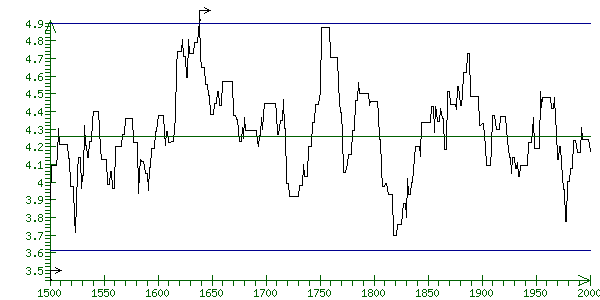
Fig. 2. An element of the profile Score(i) for searching the sequence extracted from EMBL by AC=X73839, with the located transcription start, for the promoter extracted form the TRRD database by ID=Hs:PBGD.
The results of the application of Equations 1 and 2 to a sequence extracted form EMBL by AC=X73839 (A. thaliana gene for hemC) and promoter of porphobilinogen deaminase extracted from TRRD by ID=Hs:PBGD are shown in Fig. 2. Note that the algorithm described above predicted one potential transcription start at position 1638 in this sequence. According to the information contained in the field FT, this sequence has the transcription start at position 1603.
Further development of this approach will employ the construction of Scoren(i) using homology search, weight matrices, perceptrons, and other methods. Constructions of control and test samples will be refined methodologically basing on extraction of experimental footprints, local Gibbs aligning, and construction of recognition groups.
The TRRD database itself will be integrated with the system described above through cross-hyperreferences and automatic generation of the recognition programs for a promoter described in TRRD and intended to be searched for in a user’s sequence. We also plan to supplement the system with the modules for analysis of functional site activity based on the di- and trinucleotide composition.
The work was supported by the Russian Foundation for Basic Research (97-04-90309) and Russian Human Genome Project.
References
- Etzold, T. and Argos, P. (1993) SRS–an indexing and retrieval tool for flat file data libraries. CABIOS. 9, 49-57
- Kel, A.E., Kolchanov, N.A., Kelí, O.V., Romashchenko, A.G., Ananíko, E.A., Ignatieva, E.V., Merkulova, T.I., Podkolodnaya, O.A., Stepanenko, I.L., Kochetov, A.V, Kolpakov, F.A., Podkolodny, N.L., and Naumochkin A.N. (1997) TRRD: database on transcription regulatory regions of eukaryotic genes. Mol. Biol. (Msk) 31, 521-530.
- Kel, O.V., Romashchenko, A.G., Kel, A.E., Wingender, and E., Kolchanov, N.A., (1995b) A compilation of composite regulatory elements affecting gene transcription in vertebrates. Nucl. Acids Res. 23, 4097-4103.
- N.A. Kolchanov, M.P. Ponomarenko, A.E. Kel, Y.V. Kondrakhin, A.S. Frolov, F.A. Kolpakov, O.V. Kel, E.A. Ananko, E.V. Ignatieva, O.A. Podkolodnaya, I.L. Stepanenko, T.I. Merkulova, V.N. Babenko, D.G. Voroblev, S.V. Lavyushev, Y.V. Ponomarenko, A.V. Kochetov, G.B. Kolesov, N.L. Podkolodny, L. Milanesi, E. Wingender, T. Heinemeyer, V.V. Solvyev “Genexpress: A Computer System for Description, Analysis and Recognition of Regulatory Sequences in Eukaryotic Genome” ISMB’98, in press (1998)
- Peter, R.C., Juner, T., and Bucher, P. (1998) The eukaryotic promoter database EPD. Nucl. Acids Res. 26, 353-357.
- Salas, F., Haas, J., Brunk, B., Stoeckert Jr, C.J., and Overton, G.C. (1998) EpoDB: a database of genes expressed during vertebrate erythropoiesis. Nucleic Acids Res. 26, 290-292
- Wingender, E., Dietze, P., Karas, H., and Kneuppel, R. (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucl. Acids Res. 24. P. 238-241.