KAMZOLOVA S.G.1, DZHELYADIN T.R.2+, SOROKIN A.A.1, IVANOVA N.N.1, KUTUZOVA G.I.3, ESIPOVA N.G.3, POLOZOV R.V.2
1Institute of Cell Biophysics of RAS, Pushchino Moscow region, 142292 Russia, kamzolova@uranus.iteb.serpukhov.su.
2Institute of Theoretical and Experimental Biophysics of RAS, Pushchino Moscow region, 142292 Russia, polozov@venus.iteb.serpukhov.su
3Engelhard Institute of Molecular Biology, 32 Vavilov street, Moscow, 117984, Russia.
+Corresponding author:timurd@geocities.com
Keywords: molecular mechanisms, promoter-polymerase recognition, electrostatic interactions, promoters
Electrostatic potential was calculated in accordance with Coulomb’s law for several tens of promoters recognized by E.coli RNA polymerase Es 70. Some correlation between patterns of electrostatic potential of promoters and their attributing to certain promoter classes was found. The data obtained indicate that electrostatic field can be an additional component in promoter–polymerase recognition coding, acting at the middle distances at initial stage of their interaction.
Introduction
Statistical averaging of 290 sequenced promoters recognized by E.coli RNA polymerase (Es 70) has revealed two consensus hexamers, TTGACA and TATAAT located 35 and 10 base pairs upstream from the start of transcription [1]. The conserved sequences deviate appreciably in individual promoter exhibiting an average conservation of 7.9 nucleotides per promoter in the both regions. Therefore, the consensus hexamers as universal coding sequences appeared to be of little predictive value for individual promoters and could not be used in generating of an efficient promoter-site searching algorithm. In addition, the efficiency of some promoters was found to be dependent not only on the –10 and the –35 regions but to be determined also by nucleotide sequences of some other sites. It should be emphasized that these functionally important sites differ for different promoters and their groups [reviewed in 2]. It was proposed that promoter activity could be coded by different programs, which is reflected by structural differentiation of recognizable elements for different promoter groups. In accordance with this conception the problem of promoter–polymerase recognition code should be contemplated from the standpoint of a classification analysis of promoters [2]. Some attempts have been made to develop a general approach for classification analysis of the whole set of studied promoter sequences by using the known mathematical methods such as Fourier-, wavelet-transforms and cluster analysis, neural networks and statistical weighting of individual base pairs and their combinations [3-6]. The possibility of promoter subgrouping was demonstrated in these studies. Both positive and negative correlations in the results of attributing individual promoters to particular promoter groups were observed. No general rules connecting promoter primary structure with its functional strength were formulated. Moreover it was becoming evident that other features of promoters besides their primary structure are important for promoter-polymerase recognition process and should be taken into account in the study of the problem.
Earlier it has been suggested that electrostatic forces are involved in initial binding of RNA-polymerase to DNA and identification of promoter sites [7]. In this work we studied the role of electrostatic interactions in promoter functioning. Electrostatic potentials were calculated and comparatively analyzed for several tens of E.coli promoters
Systems and methods
The next set of promoters was chosen for analysis [1]:
- the group of promoters of T5 phage — PD/E20 , PH207 , PN25, PG25, PJ5;
- promoters of T7 phage — PA1, PA2, PA3, PD;
- PL belonging to l phage;
- artificial promoters — PtacI, Pcon, Pbla ;
- E.coli promoters — Plac, PlacUV5, Ptrp, Pace, rrnBP1, rrnBP1(-UP), galP1, galP2, and some others.
All sequences were aligned in accordance with the start point of transcription.
The choice of promoters was dictated by their belonging to certain classes of phage or E.coli promoters with well characterized recognizable elements. The availability of biochemical and genetic data for these promoters was also taken into account to have an opportunity of comparing the results of electrostatic calculations with promoter activity.
Nonpromoter sequences from coding regions of E.coli recA gene were added to the set as a control system.
DNA molecule was described in terms of its three-dimensional structure as determined by the coordinates of all its atoms. The coordinates of the idealized form of B-DNA were generated from the local coordinate of Chandrasekaran and Arnott [8] by adding multiples of 36° to twist angle and the same multiples of 3.38A along helix axis per base pair.
Calculations of electrostatic potential V were carried out in vacuum in accordance with Coulomb law:
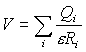 ,
,
where RI – is a distance from atom with the charge Qi to the point of consideration, e – dielectric constant. Different approximations that are used for the calculation of electrostatic properties of DNA molecule were reviewed in [9]. It should be emphasized that more accurate calculation, like Poisson-Boltzmann or Debye-Huckel can be carried out only for short fragments of DNA or periodic structure. However, the topology of electrostatic potential calculated for polyd(AT)· polyd(AT) and other periodic sequences in accordance with Debye-Huckel model in [10] was found to be in good agreement with our results obtained by simple coulombic calculations. Since the simple electrostatic calculation keeps the main features of potential distribution, it is quite suitable for qualitative analysis in comparative studies and thus can be used for the purpose of our work.
Potential was calculated on the surface of the cylinder around promoter with radius of 15A. In this case the surface of calculations is situated at about 5A from DNA sugar-phosphate backbone. The zero axis for rotation angle (twist angle) was given by the direction to the center of the first pair. The step along helix axis was equal to 1A and the step of 1° was chosen for the twist angle. Charges were assigned to the center of each atom. The values of the charges were taken from [11].
The distribution of electrostatic potential was presented as topological map.
Results and Discussion
Calculation of electrostatic potential was made for all promoters in the chosen set. Examples of nucleotide sequences of some promoters and topological patterns of their electrostatic potentials are given in figure 1. Electrostatic potential calculated for nonpromoter sequence from coding region of E.coli recA gene is also presented.
It can be seen that the patterns of electrostatic potentials of promoter and nonpromoter sequences essentially differ. Topology of electrostatic potential of nonpromoter region is more monotonous. Alternating bands of positive and negative potentials tend to be homogeneous along the helix axis. Topological maps of promoters are much less regular and more rich in electrostatic details.
Comparative analysis of several tens of promoter electrostatic replicas revealed their considerably varying individual character. However some common features could be observed in promoters belonging to groups with identical recognizable elements, such as PH207 and PG25 from the group of T5 phage promoters [12] in figure 1(b,c). It is known that this group of promoters is characterized by four promoter determinants which are located at –45, –35, -10, +7. Both promoters contain identical consensus sequences in –45 and –10 regions, their –35 consensus differ by one nucleotide; however PG25 in contrast to PH207 has no recognizable element in +7 region. In spite of this difference, they interact with RNA polymerase in analogous way as judged by their belonging to the same promoter class based on biochemical and genetic data [12]. It should be noted that this classification is in agreement with our results of electrostatic analysis of these promoters: their electrostatic potentials are quite similar in general distribution of hills and valleys, although their electrostatic patterns differ in finest details (figure 1 b,c). These data may indicate that the initial recognition of promoters by RNA polymerase is possibly controlled at the middle distances by electrostatic forces.
To compare the behavior of electrostatic potential of promoters belonging to different promoter classes, two promoters – rrnBP1 and PH207 were chosen (figure 1b, d). It should be noted that rrnBP1 and PH207 have two identical consensus sequences in -10 and -35 regions. Despite the similarity in these recognizable regions the promoters were identified as belonging to different classes on the basis of genetic and biochemical data [13]. Their attribution to different classes is due to the presence of powerful third promoter determinant (so called «UP element») in rrnBP1 promoter [13] and its absence in PH207 [12].The figure 1 (b, d) represents the distribution of electrostatic potential of these promoters. It is interesting that the potential pictures are quite different for these two promoters. This result is in agreement with their attributing to different classes.
Concluding remarks
Comparative analysis of electrostatic potentials calculated for several tens of promoters indicates that electrostatic field can be an additional component in promoter–polymerase recognition coding acting at the middle distances at initial stage of their interaction.
Figure 1 Representation of electrostatic potentials from:
a) coding sequence of E.coli recA gene (first 100 base pair),
agagaagcctgtcggcaccgtctggtttgcttttgccactgcccgcggtgaaggcattacccggcgggatgcttcagcggcgaccgtgatgcggtgcgtc
b) PH207 promoter from –51 to +19 bp,
ttttaaaaaattcatttgctaaacgcttcaaattctcgtataatatacttcAtaaattgataaacaaaaa
c) PG25 promoter from –50 to +20 bp,
tgaaaaataaaattcttgataaaattttccaatactattataatattgttAttaaagaggagaaattaac
d) rrnBP1 promoter from –61 to +1 bp
tcagaaaattattttaaatttcctcttgtcaggccggaataactccctataatgcgccaccA.
X axis–dinucleotide position along the promoter sequence, Y axis–a rotation angle. Color scale–an electrostatic potential in units of charge of proton per angstrom.
References
- Sh.Lisser and H. Margalit, «Compilation of E.coli mRNA promoter sequences» Nucl. Acids Res. 21, 1507 (1993)
- S.G. Kamzolova, «Interaction of Escherichia coli RNA polymerase with promoters. The necessity for a classification approach to the problem of promoter–polymerase recognition» Biochemistry (Moscow) 60, 285 (1995)
- G.I. Kutuzova, G.K. Frank, V.Yu. Makeev, N.G. Esipova and R.V. Polozov, «Fourier analysis of nucleotide sequences. Periodicities in E.coli promoter sequences» Biofizika 42, 354 (1997)
- F. Roskot, P. Sazelova and L.Pivec, «A novel method for promoter search enhanced by function–specific subgrouping of promoters—developed and tested on E.coli system» Nucl. Acids Res. 17, 4799 (1989)
- A.V. Lukashin, V.V. Anshelevich, B.R. Amirikyan, A.J. Gragerov and M.G. Frank-Kamenetskii, «Neural network model for promoter recognition» J. Biomolec. Struct. Dynamics 6, 1123 (1989)
- K. Weller and R.-D. Recknagel, «Promoter strength prediction based on occurence frequences of consensus patterns» J. Theor. Biol. 171, 355 (1994)
- J.D. Gralla, «Promoter recognition and mRNA initiation by Escherichia coli» Methods in Enzymology. Gene Expression Technology (D.V. Goeddel ed., Academic Press, San Diego, California, 1991)
- S. Arnott, P.J.C. Smith, R. Chandrasekaran, In. «CRC Handbook Biochemistry and Molecular biology: Nucleic Acids» 2,(G.D. Fasman, CRC Press, 1976)
- B. Jayaram, K. A. Sharp and B. Honig, «The Electrostatic Potential of B-DNA» Biopolymers 28, 975 (1989)
- K. Wagner, E. Keyes, T. W. Kephart and G. Edwards, «Analytical Debye–Huckel Model for electrostatic potentials around dissolved DNA» Biophys. J. 73, 21 (1997)
- V.I. Poltev, A.V. Teplukhin, G.G. Malenkov, «Computational Investigation of the Role of Hydration in Nucleic Acid Structure and Function» Int. J. Quant. Chem. 42, 1499 (1992)
- U. Deuschle, W. Kammerer, R. Gentz and H. Bujard, «Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures» EMBO J. 5, 2987 (1986)
- W. Ross, K. K. Gosink, Y. Solomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov and R. L. Gourse, «A third recognition element in bacterial promoters: DNA binding by the a subunit of RNA polymerase» Science 262, 1407 (1993)

