TAKANORI WASHIO1, TOMITA MASARU2
Laboratory for Bioinformatics
1Graduate School of Media and Governance, e-mail: washy@sfc.keio.ac.jp
2Department of Environmental Information; e-mail: mt@sfc.keio.ac.jp
Keio University, 5322 Endo, Fujisawa, 252 Japan
1. Introduction
Hairpin loop structures, formed in mRNAs of E. coli, are believed to be involved with transcription termination [1]-[9]. There are two different mechanisms for transcription termination in E. coli: rho-dependent and rho-independent termination. In rho-independent termination, the elongation process of RNA polymerase is believed to be terminated by a hairpin loop in the tertiary structure of mRNA, while in the rho-factor dependent process transcription is terminated by the interaction of the rho factor protein with RNA polymerase. Many researchers think that this is the general mechanism of procaryotic transcription termination.
In the present study, we systematically computed the free energy values of mRNA tertiary structures around stop codons in order to surmise the hairpin-forming potential for all genes in each of the eleven complete procaryote genomes– E. coli K-12 [15], H. influenzae Rd [10], B. subtilis [17], Mycoplasma genitalium [11], Mycoplasma pneumoniae [12], Synechocystis PCC6803 [13], Helicobacter pylori [16], Borrelia burgdorferi [20], Methanococcus jannaschii [14], Archaeoglobus fulgidus [18], and Methanobacterium thermoautotrophicum [19]–.
Methods
The free energy values were calculated using the RNA tertiary structure prediction program, “RNAfold”, developed by Hofacker et al [21]. The “RNAfold” program applied the algorithm used in a program called “mfold” [22]. We set the window gauge at 60 bp, and the window is shifted by 10 bp. A nucleotide sequence length of 60 bp is, thus, fed to the RNAfold program each time the window is shifted. Putative free energy values are computed from 200 bp upstream to 200 bp downstream of each stop codon. The average free-energy values at each point around the stop codon over all ORFs were then plotted.
Results and discussion
In procaryotes, several genes in a specific pathway are often organized into an operon, which is transcribed into a polycistronic mRNA. Those genes within an operon, except the last gene, are not involved in transcription termination. Since reliable operon prediction from genome sequence is not possible, we are unable to reliably distinguish genes with termination sites from those without. However, we know that regions between two “end-on” genes with opposite directions must contain transcription termination sites for these genes. We classified all intergenic regions into the following three categories: The direction of two adjacent genes can be the same direction (– > –>), “end-on” (–> <–), or “head-on” (<– –>). While transcription termination sites must always exist between “end-on” genes, there may or may not exist a termination site between genes in the same direction, since they may be a part of an operon structure. Termination sites usually do not exist between “head-on” genes.
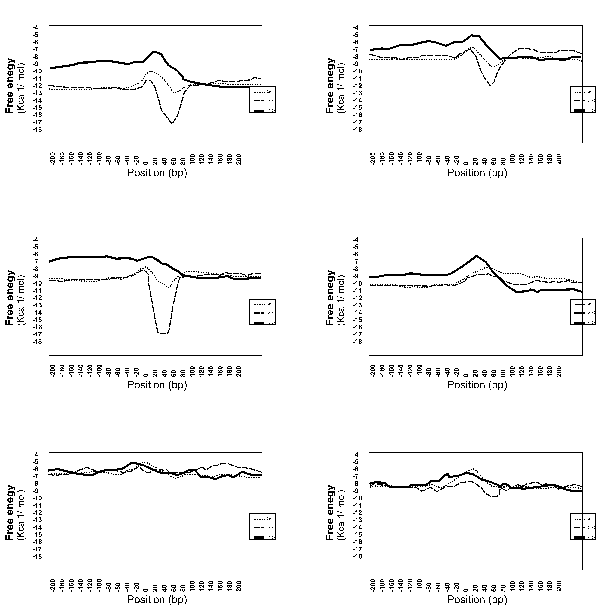
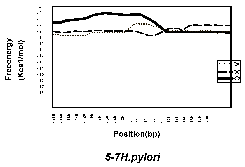
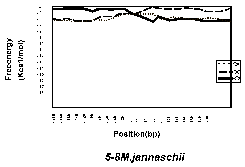
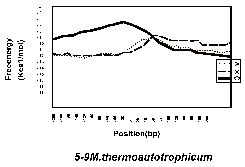
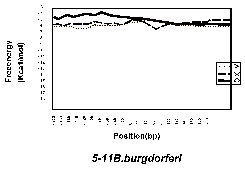
Figure 1 shows the free-energy values around stop codons for each of the three orientations of adjacent genes. In E. coli K-12, H. influenzae Rd, and B. subtilis, we observed a sharp drop in free-energy values between “end-on” genes, but much less between genes in the same direction, and no steep drop is found between “head-on” genes. Those results validate our assumption that these free-energy drops indeed represent hairpin-forming sequences at transcription termination sites. On the other hand, eight other procaryotes– Mycoplasma genitalium, Mycoplasma pneumoniae, Synechocystis PCC6803, Helicobacter pylori, Borrelia burgdorferi, Methanococcus jannaschii, Archaeoglobus fulgidus, and Methanobacterium thermoautotrophicum— had showed no apparent decrease in free energy value at the corresponding regions. This result suggests that these procaryotes, or at least some of them, may never form hairpins at their transcription termination sites.Finally, it should be noted that the lack of apparent free-energy drop does not necessarily indicate the complete absence of hairpins. However, their involvement with transcription termination, if any, is probably different qualitatively from that of the species with apparent hairpin formation. We therefore conclude that the well-understood mechanism of transcription termination of E. coli involving hairpin formation is not a universal mechanism of procaryotes.
Fig. 1. Free enery values around stop codons foreach of the three orientations of adjacent genes
 |
 |
 |
 |
 |
Acknowledgments
We are deeply grateful to Hiroji Aiba and Hiroyuki Abe (Nagoya University, Nagoya, Japan), Hiroyuki Mori, Takeshi Ito and Naotake Ogasawara (Nara Graduate School of Advanced Science and Technology, Nara, Japan), Takakazu Kaneko (Kazusa DNA Institute, Kisarazu, Japan), and Hidemi Watanabe (National Center of Biotechnology Information, Maryland , U.S.A.) for valuable suggestions on the bacterial transcriptional termination systems. We also thank Rintaro Saito, Ryo Matsushima and Takuya Sakaguchi (Keio University, Kanagawa, Japan) for their help in computer programming, and Tom Shimizu for his comments on the early draft of this manuscript.
This work is supported in part by a Grant-in-Aid for Scientific Research on Priority Areas ‘Genome Science’ from The Ministry of Education, Science, Sports and Culture in Japan.
References
- Roberts Roberts, J. W. (1969) Nature, 224(225), 1168-74.
- Adhya, S. and Gottesman, M. (1978) Annu Rev Biochem, 47, 967-96.
- Platt, T. (1981) Cell, 24(1), 10-23.
- Platt, T. (1986) Annu Rev Biochem, 55, 339-72.
- Friedman, D. I., Imperiale, M. J. and Adhya, S. L. (1987) Annu Rev Genet, 21, 453-88.
- Das, A. (1993) Annu Rev Biochem, 62, 893-930.
- Reynolds, R. and Chamberlin, M. J. (1992) J Mol Biol, 224(1), 53-63.
- Reynolds, R., Bermudez-Cruz, R. M. and Chamberlin, M. J. (1992) J Mol Biol, 224(1), 31-51.
- d’Aubenton Carafa, Y., Brody, E. and Thermes, C. (1990) J Mol Biol, 216(4), 835-58.
- Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty,B. A., Merrick, J. M. and et al. (1995) Science, 269(5223), 496-512.
- Fraser, C. M., Gocayne, J. D., White, O., Adams, M. D., Clayton, R. A., Fleischmann, R. D., Bult, C. J., Kerlavage, A. R., Sutton, G.,Kelley, J. M. and et al. (1995) Science, 270(5235), 397-403.
- Himmelreich, R., Hilbert, H., Plagens, H., Pirkl, E., Li, B. C. and Herrmann, R. (1996) Nucleic Acids Res, 24(22), 4420-49.
- Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S.,Kimura, T., Hosouchi, T., Matsuno, A., Muraki, A., and et al. (1996) DNA Res, 3(3), 109-36.
- Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., Blake, J. A., FitzGerald, L. M., Clayton, R. A.,Gocayne, J. D., Kerlavage, A. R., Dougherty, B. A., Tomb, J. F. and et al. (1996)Science, 273(5278), 1058-73.
- Blattner, F. R., Plunkett, G., 3rd, Bloch, C. A., Perna,N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D.,Rode, C. K., Mayhew, G. F., Gregor, J., Davis, N. W., Kirkpatrick,H. A., Goeden, M. A., Rose, D. J., Mau, B. and Shao, Y. (1997)Science, 277(5331), 1453-74.
- Tomb, J. F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., Nelson, K., Quackenbush, J., Zhou, L. and et al. (1997) Nature, 388(6642) 539-47.
- Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessieres, P., Bolotin, A., Borchert, S., Borriss, R., Boursier, L., Brans, A., Braun, M., Brignell. and et al. (1997) Nature, 390(6657), 249-56.
- Klenk, H. P., Clayton, R. A., Tomb, J. F., White, O., Nelson, K. E., Ketchum, K. A., Dodson, R. J., Gwinn, M., Hickey, E. K., Peterson, J. D., Richardson, D. L., Kerlavage, A. R., Graham, D. E. and et al. (1997) Nature, 390(6658), 364-70.
- Smith, D. R., Doucette-Stamm, L. A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D., Cook, R., Gilbert, K., Harrison, D., Hoang, L., Keagle, P., Lumm, W., Pothier, B., Qiu, D., Spadafora, R., Vicaire, R., Wang, Y., Wierzbowski, J., Gibson, R., Jiwani, N., Caruso, A., Bush, D., Reeve, J. N. and et al. (1997) J Bacteriol, 179(22), 7135-55.
- Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., White, O., Ketchum, K. A., Dodson, R., Hickey, E. K., Gwinn, M., Dougherty, B., Tomb, J. F. and et al. (1997) Nature, 390(6660), 580-6.
- I.L. Hofacker, W. Fontana, P.F. Stadler,S. Bonherffer, M. Tacker, P. Schuster. (1994) Monatshefte f. Chemie125, 167-188.
- Zuker, M. and Stiegler, P. (1981) Nucleic Acids Res, 9(1), 133-48.