SHAHMURADOV I.A.+, MUSTAFAYEV N.Sh., ALIEV J.A.
Institute of Botany, Azerbaijan Academy of Sciences, Baku 370073, Azerbaijan Republic; e-mail: dj_aliev@azeri.com
+Corresponding author
Keywords: transcription regulation, chloroplasts, regulatory elements, nukleotide sequences
1. Introduction
Genetic regulation of biogenesis and function of chloroplasts is carried out co-operatively by nuclear and plastid genomes. It includes coordinated expression of chloroplast and some nuclear genes. Regulation of these genes occurs at various levels (transcription, post-transcription and translation), but the first level of regulation is transcriptional, where light is a initial signal for induction, at least, of some genes transcription. Regulation of transcription is carried out by various regulatory elements (RE), interacting with corresponding transcription factors. As yet it is unknown regulatory elements of such coordinated gene transcription. Although regulatory mechanisms of transcription of chloroplast and nuclear genes essentially differ, but it may be prove extremely interesting if the coordinated regulation and influence of same environmental factors to both genomes is an indication of existence of some common elements of transcription regulation. Furthermore, it is quite possible that there are some other RE exclusively related to chloroplast genes. Therefore, to detect potential regulatory elements of chloroplast genes a transcription computer analysis of upstream promoter regions of all 113 identified chloroplast genes/ORFs from Oryza sativa, as model system, with the exception of tRNA genes, has been performed. Data obtained indicate that, at least, some of chloroplast genes perhaps contain functional RE resembling those in nuclear genes. Moreover, it is not ruled out that there are some other RE exclusively related to chloroplast genes. Here we present some of the results obtained.
2. Materials and Methods
DNA sequences of complete chloroplast genomes of rice and tobacco (for comparative analysis) were extracted from the GenBank database. The sequences of upstream regions of all 113 analyzed chloroplast genes are 1050 bp long and contain 1000 bp upstream and 50 downstream of translation start point (coordinates – 1000: +50; here and everywhere + 1 indicate the first nucleotide of translation start codon). To search for DNA motifs homologous to known transcription RE DNA sequences of 120 RE from various plant nuclear genes, 3 Light Response Element (LRE) of tobacco chloroplast psbD gene and 2 mammalian RE resembling corresponding RE of plants (carbohydrate response elements) have been used. A search for putative RE was carried out by “SITE” computer method (Shahmuradov et al., 1986). A comparative analysis of promoter regions of corresponding genes of rice and tobacco was carried out by “FASTA” (Pearson, 1990) and “GOMOL” (Kolchanov et al., 1985) program packages.
3. Results and Discussion
5í-promoter regions (- 1000: +50) of 113 rice chloroplast genes/ORFs and some of corresponding genes of tobacco have been analyzed by all three methods above mentioned. Although the analysis didnít reveal common RE for all analyzed genes (with exception of canonical -35 and -10 boxes), but areas of extensive and statistically nonrandom homology between promoter sequences of chloroplast genes of various groups have been found (Fig. 1-3). Moreover, most of these motifs are frequently presented by the several copies in the same promoter region (data not shown). Some of these motifs are similar to various known nuclear REs. Thus, for example, a part of the motif M5 in inverted orientation (aTTCAAA) is similar to the LRE1 (TTTCAAA) of pea rbcS-3A gene and motif M6 contains sequence (TGGTTT) identical to anaerobic responsive element of maize adh1, tomato adh2 and ldh1 genes (Fig. 1). Furthermore, some of these common elements are also present in the promoter regions of corresponding chloroplast genes of tobacco and, consequently, are conservative (Fig. 2).
Figure 1. Some of DNA motifs found in the promoter regions, at least, of 5 various rice chloroplast genes.
Figure 2. Homologous DNA motifs found in the promoter regions of rice and tobacco chloroplast psbD and rbcL genes. Conservative sequence ATGG found in the promoters of various chloroplast genes underlined. DRE ñ drought response element of Arabidopsis thaliana.
Besides, the analysis also revealed that a set of various DNA motifs resembling the known REs of the nuclear and chloroplast genes are present in the upstream regions of analyzed chlo-roplast genes (Fig. 4). Interestingly, promoter sequences of corresponding tobacco genes also contain some of these putative RE (data not shown). One of the most notable features of all promoter sequences analyzed is the presence of motifs similar to known plant nuclear RE, responsive to various factors (light, heat-shock, sucrose abundance, anaerobic stress, drought stress, metal ions, nitrate, salicylic acid, etc.) or hormones (ethylene, auxin). Thus, it is a well established fact that some nuclear genes encoding proteins involved in photosynthesis are
Figure 3. Homologous DNA motifs found in the promoter regions of rice psbA gene and some other rice chloroplast genes.
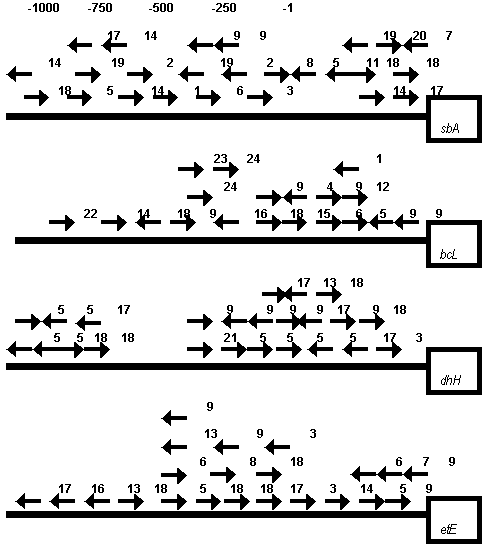
Figure 4. DNA motifs homologous to known transcription RE in the upstream promoter regions of rice chloroplast encoded psbA, rbcL, ndhH and petE genes. Motifs on the coding and complementary DNA strands are denoted by corresponding arrows under the line. The following conventional signs have been used: 1 – accessory factor binding site (mammalian S14 gene promoter); 2 – SP8 binding site (sweet potato sporamin A gene promoter; 3 – sucrose response element of potato patatin gene promoter); 4 – synthetic auxin response element of soybean GH3 gene; 5 – regulatory element in the pea PsMTA MT–like gene promoter; 6 ñ LRE1 of pea rbcS-3A gene; 7 – LRE3 of pea rbcS-3A gene; 8 – LRE3* of pea rbcS-3A gene; 9 – LRE_A1/A2 of tobacco psbD gene; 10 – ethylene-responsive element in promoters of defence-related genes; 11 – HSE of pea hsp70 gene; 12 – HSE of soybean HSE 6871 gene; 13 – seed-specific enhancer element of Arabidopsis AtPer1 gene; 14 – anareobic responsive element of maize adh1, tomato adh2 and ldhl genes; 15 – conservative element in the promoter region of rice rbcL, psbD and some other chloroplast genes (see Fig.2, b ñ c); 16 – root specific element of some plant genes; 17 – Myb-binding site; 18 – Myc-binding site; 19 – unknown element found in the promoter regions of some plant genes; 20 – GT-1 binding site; 21 – GATA box of petunia cab22R gene; 22 – regulatory element (GRA) in the promoter of ABA responsive gene from maize; 23 – SURE of Phaseolus vulgaris rbcS2 gene (see also text); 24 ñ SURE of potato patatin gene (see also text).
subject to negative feedback regulation by sugars, accumulated through photosynthetic carbon fixation. To present a number of various sucrose response (repression or induction) elements (SURE) from several nuclear genes, including rbcS, have been identified (Urvin, Jenkins, 1997). Our analysis revealed a presence of two motifs (-590: TTcCaCATGG; -470: tATACTAcT) in the upstream region of rice rbcL gene, similar to SUREs of Phaseolus vulgaris rbcS2 gene (TTACTCATGG) and potato patatin genes ( AATACTAAT), respectively (see also Fig. 4). It is particularly interesting in connection with necessity of co-ordinated expression of rbcS and rbcL genes, coding small and large subunits of ribulose 1,5-bisphosphate carboxy-lase/oxygenase, involved in photosynthesis.
Moreover, a set of motifs resembling (with one mismatch) the regulatory site (ATTCAAA) found in the promoter region of the pea metallothionein (MT)-like gene PsMTA are present in the upstream regions of genes, coding different subunits of NADH dehydrogenase. Thus, each of the genes coding NADH dehydrogenase subunit 3 (ndh C) and NADH dehydrogenase subunit 4 (ndh D), respectively, contains such 4 motifs (data not shown), and the gene coding NADH dehydrogenase 49 kDa protein (ndh H) contains even 8 motifs (see Fig. 4). Nuclear genes encoding predicted plant proteins with sequence similarity to MTs were identified in a wide range of various species. There is circumstantial evidence to suggest that the products of these genes could have roles in trace metal ion homeostasis (Fordham-Skelton et al., 1997). Such motifs also found in the promoters of some other chloroplast genes analyzed by us. A possible role of these abundant motifs and other conservative sequences/putative RE found in various chloroplast genes remains to be elucidated.
4. Conclusion
Our results indicate that at least some of chloroplast genes perhaps contain functional RE similar to those in nuclear genes. Moreover, it is not ruled out that there are some other RE exclusively related to chloroplast genes.
References
- Fordham-Skelton A.P., Lilley C., Urwin P.E., Robinson N.J. GUS expression in Arabidopsis directed by 5í regions of the pea metallothionein gene PsMTA. Plant Mol.Biology, 34, 659 (1997).
- Kolchanov N.A., Solovyov V.V., Zharkikh A.A. Contextual analysis of polynucleotide sequen-ces. Methods of search for nonrandom repeats. Molecular Biology (in Russian), 19, 524, (1985.)
- Pearson W.R. (1990) Rapid and Sensitive Sequence Comparision with FASTP and FASTA. Methods in Enzymology, 183, 63.
- Shahmuradov I.A., Kolchanov N.A., Solovyov V.V., Ratner V.A. Enhancer-like structures in middle repetitive DNA elements of eukaryotic genomes. ñ Genetika (in Russian), 22, 357, 1986.
- Urvin N.A.R., Jenkins G.I. A sucrose repression element in the Phaseolus vulgaris rbcS2 gene promoter resembles elements responsible for sugar stimulation of plant and mammalian genes. Plant Mol.Biology, 35, 929 (1997).