Novosibirsk State University, Novosibirsk, 630090 Russia;
Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 10 Lavrentiev Ave., Novosibirsk, 630090, Russia
Keywords: functional motifs, mobile genetics elements, molecular functions
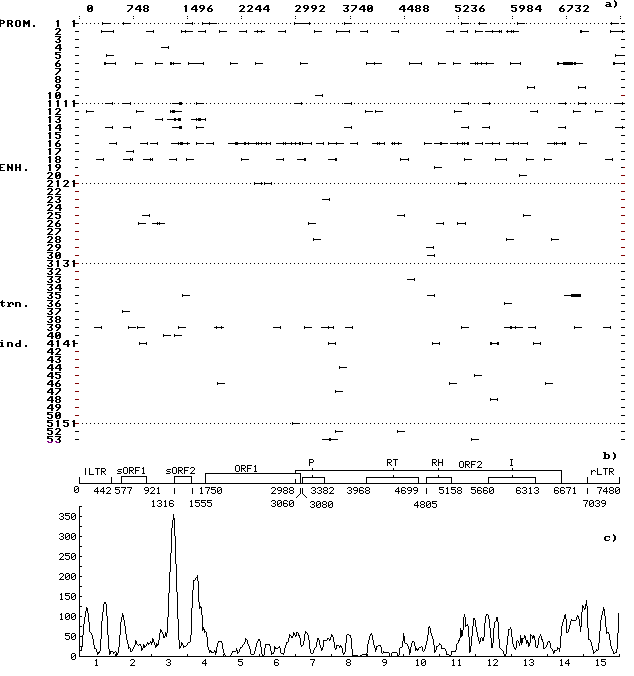
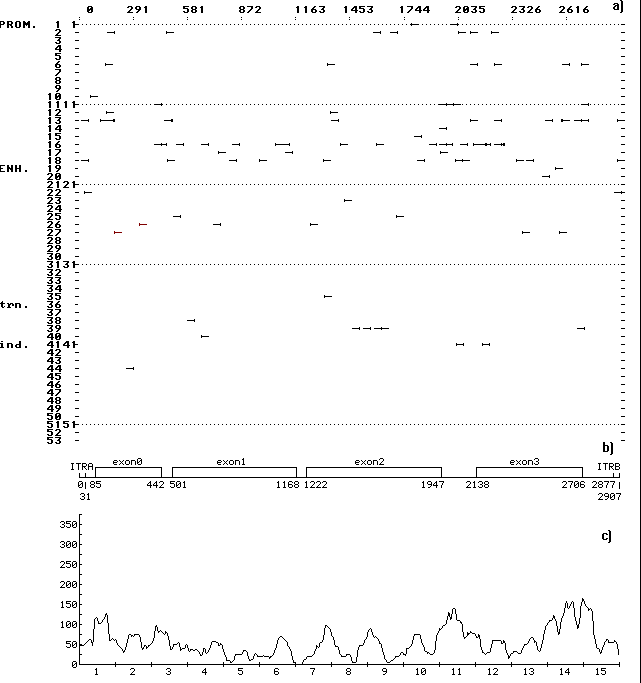
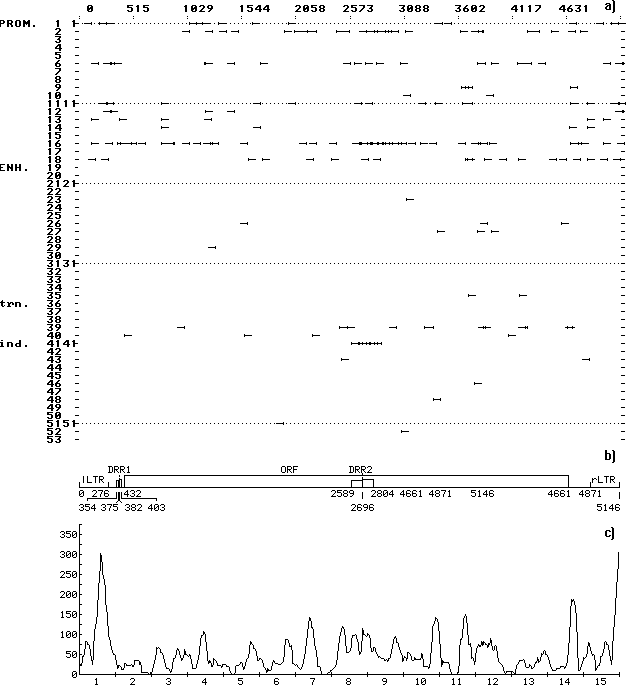
A computer-assisted analysis of DNA sequences of 19 mobile genetic elements (MGE) of Drosophila and other objects (MDG1, MDG2, MDG4, 17.6, 297, LINE1, etc.) was performed. Over 500 motifs of 80-90 types that are similar to known functional sites compiled in the database (comprising 278 units) were revealed in each case. The motifs were shown to be unevenly distributed along the DNA sequence: the correlation between the regions of motif “jamming” and the locations of potential regulatory regions is highly significant. For example, the sequence of Drosophila retrotransposon MDG1 (Fig. 1) contains the highest maximums (“jams”) in segments 1 and 15 (within LTR), 2 (in the region close to sORF1 left terminus), 3 (the interval the interval between sOFR1 and sORF2), 4 (the interval between sOFR2 and ORF1), 11 and 12 (the interval between RH and I), 14 and 15 (the interval between ORF2 and rLTR). In transposon P of Drosophila (Fig. 2), the “jams” were revealed in segments 1 (the interval between ITRA and exon 0), 11 (the interval between exon 2 and exon 3), 14 (the region close to the right terminus of exon 3), and 15 (the interval between exon 3 and ITRB). The highest maximums of functional site distribution in the Drosophila copia element (Fig. 3) are located in segments 1 and 15 (within LTR), 8 (the region close to the left end of DRR2 and inside DRR2) and 14 (the interval between ORF and rLTR). Being located in these positions, the motifs of functional sites can provide the regulation of basic MGE functions: expression of their own ORFs, reproduction (transposition), transposition induction, modifying effect on neighboring genes and polygenes, etc. However, several dozens of functional sites are sufficient to provide for the MGE molecular functions. Their overabundance suggests the role of MGE as “mobile cassettes of functional sites” with an increased content of “semi-finished” motifs that could be easily transformed into functional sites.
Statistical significance of these jams was proved by comparing with random sequences of the same lengths and nucleotide compositions as the MGE MDG1, MDG2, MDG4, 17.6, 279, LINE1, etc., analyzed. Distribution of the motifs correlates significantly with the distribution of nucleotide composition (%A+T).
The work was supported by the Russian Foundation for Basic Research (grant No. 97-04-49232), Program for Soros Professorship (grant No. 683p), and the program of the Russian Ministry of Education “Universities of Russia: Basic Research”.
Fig. 1. (a) Distribution of the revealed motifs of functional sites along the MDG1 DNA sequence in the individual samples. The ascribed numbers of the regulatory sites are indicated on the ordinate; arrows mark the discovered location of regulatory sites on the left-directed (leftward arrows) and right-directed (rightward arrows) DNA strands; prom, the sites of replication and transcription initiation and termination; enh, enhancers and silencers of chromosomal, viral, etc., genes; trn, the sites recognized by cellular protein transcription and translation factors; and ind, the sites recognized by protein receptors for inductive signals.
(b) Schematic structure of MDG1 DNA sequence: LTR, long terminal repeats; sORF1 and sORF2, small open reading frames; ORF1 and ORF2, big open reading frames; P is the motif of amino acid sequence of protease domain; RT, of reverse transcriptase domain; RH, of RNase H domain; and I, of integrase domain.
(c) Consolidated distribution of the revealed motifs of functional sites along the MDG1 DNA sequence. Scanning was performed with a window 75 nt long and scanning span of 15 nt. Numbers of segments are indicated on the abscissa; total number of nucleotides contained in the motifs of functional sites and falling within the window, on the ordinate. Jamming of the motifs correlates with the potential regulatory regions in the LTR and in the vicinity of the ends (start and beginning) of small and big ORFs and domains of the big ORF2.
Fig. 2. (a) Distribution of the revealed motifs of functional sites along the DNA sequence of Drosophila transposon P in the individual samples. The designations are as in Fig. 1.
(b) Schematic structure of the DNA sequence of transposon P: ITRA, left inverted tandem repeat; ITRB, right inverted tandem repeat; the rest designations are as in Fig. 1.
(c) Consolidated distribution of the revealed motifs of functional sites along the DNA sequence transposon P. The designations are as in Fig. 1. Jamming of the motifs correlates with the locations of potential regulatory regions in the intervals between exons and between exon 3 and rLTR.
Fig. 3. (a) Distribution of the revealed motifs of functional sites along the DNA sequence of Drosophila copia element in the individual samples. The designations are as in Fig. 1.
(b) Schematic structure of the DNA sequence of Drosophila copia element: DRR1 and DRR2, direct repeats; the rest designations are as in Fig. 1.
(c) Consolidated distribution of the revealed motifs of functional sites along the DNA sequence of Drosophila copia element. The designations are as in Fig. 1. Jamming of the motifs correlates with the locations of potential regulatory regions within LTR, DRR2, and in the interval between the ORF and rLTR.