Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 10 Lavrentiev Ave., Novosibirsk, 630090, Russia
Keywords: eukaryotic transcription factors, conformational and physico-chemical properties of DNA, classification
Local nonuniformity of DNA conformational and physico-chemical properties and their dependence on nucleotide content have been demonstrated in numerous studies [1, 2]. X-ray structure analysis of B-DNA duplexes and complexes with proteins provided for determination of the mean conformational parameters of each dinucleotide [3-5]. Mean values of a number of physico-chemical properties have been determined for each dinucleotide too [6-10].
A method for classification of transcription factors based on significant context-dependent B-DNA conformational and physico-chemical properties of their binding sites, revealed by the system B-DNA-VIDEO [11] is proposed in this work. Sets of nucleotide sequences of transcription factor binding sites from the SAMPLES database without 100% homologues [12] (Table 1) were used for the analysis. The sequences in each set are aligned relative to the center of the experimentally determined binding sites of a definite transcription factor; their location was determined according to the information contained in the TRANSFAC database and published data (in case of YY1 site, the TRRD database, v 3.5, was also employed). The sequences considered was 120 bp long with flanks of 60 bp from the center of the site.
Table 1. Eukaryotic transcription factor binding sites involved in the analysis
|
Factor |
Class according to TRANSFAC |
Number of sequences in |
Size of footprint or TRANSFAC sequence element, bp |
||||
|
Number |
Name |
set |
Mean |
Min |
Max |
||
| 1 | AP-1 |
1.1 |
Leucine zipper factors (bZIP) |
69 |
14.9 |
6 |
53 |
| 2 | c-Fos |
1.1 |
– “ – |
19 |
13.9 |
6 |
29 |
| 3 | c-Jun |
1.1 |
– “ – |
28 |
14.6 |
6 |
43 |
| 4 | NF-E2 |
1.1 |
– “ – |
12 |
10.7 |
3 |
20 |
| 5 | CRE-BP1 |
1.1 |
– “ – |
22 |
12.7 |
6 |
25 |
| 6 | ATF |
1.1 |
– “ – |
25 |
12.9 |
5 |
25 |
| 7 | CREB |
1.1 |
– “ – |
37 |
13.6 |
4 |
37 |
| 8 | C/EBP |
1.1 |
– “ – |
108 |
19.1 |
4 |
96 |
| 9 | NF-IL6 |
1.1 |
– “ – |
21 |
19.9 |
8 |
41 |
| 10 | MyoD |
1.2 |
Helix-loop-helix factors (bHLH) |
16 |
18.5 |
8 |
59 |
| 11 | E2F |
1.3 |
(bHLH-ZIP) |
9 |
13.2 |
7 |
30 |
| 12 | USF |
1.3 |
– “ – |
25 |
14.1 |
5 |
30 |
| 13 | NF-1 |
1.4 |
NF-1 |
101 |
17.2 |
4 |
53 |
| 14 | RF-X |
1.5 |
RF-X |
12 |
17.0 |
11 |
24 |
| 15 | CP1 |
1.6 |
Heteromeric CCAAT factors |
33 |
17.2 |
4 |
41 |
| 16 | ER |
2.1 |
Cys4 zinc finger of nuclear receptor type |
25 |
17.5 |
4 |
40 |
| 17 | GR |
2.1 |
– “ – |
54 |
12.6 |
4 |
39 |
| 18 | PR |
2.1 |
– “ – |
20 |
11.7 |
5 |
20 |
| 19 | RAR |
2.1 |
– “ – |
16 |
24.6 |
7 |
56 |
| 20 | RXR |
2.1 |
– “ – |
21 |
20.9 |
10 |
28 |
| 21 | T3R |
2.1 |
– “ – |
21 |
20.5 |
10 |
28 |
| 22 | COUP |
2.1 |
– “ – |
17 |
20.6 |
11 |
35 |
| 23 | GATA-1 |
2.2 |
Diverse Cys4 zinc fingers |
76 |
16.1 |
5 |
41 |
| 24 | Sp1 |
2.3 |
Cys2His2 zinc finger domain |
176 |
13.1 |
4 |
96 |
| 25 | YY1 |
2.3 |
– “ – |
27 |
13.6 |
6 |
27 |
| 26 | GAL4 |
2.4 |
Cys6 cysteine-zinc cluster |
16 |
14.7 |
3 |
22 |
| 27 | EN |
3.1 |
Homeodomain |
12 |
11.4 |
6 |
20 |
| 28 | HNF-1 |
3.1 |
– “ – |
38 |
21.0 |
5 |
96 |
| 29 | OCT |
3.1 |
– “ – |
73 |
14.9 |
7 |
46 |
| 30 | HNF-3 |
3.3 |
Fork head / winged helix |
10 |
24.6 |
8 |
96 |
| 31 | c-Myb |
3.5 |
Tryptophan clusters |
19 |
18.9 |
5 |
31 |
| 32 | Ets |
3.5 |
– “ – |
15 |
20.9 |
9 |
45 |
| 33 | IRF-1 |
3.5 |
– “ – |
7 |
16.1 |
5 |
22 |
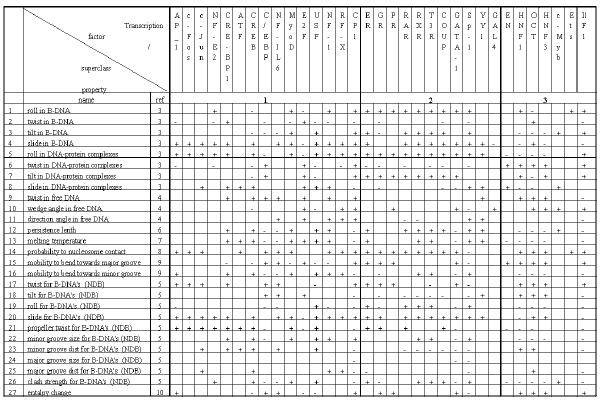
Table2. Significant conformational and physico-chemical properties of the DNA transcription factor binding sites.
Listed in Table 2 are the conformational and physico-chemical properties from the PROPERTY database used in this work. Basing on these data, the computer system B-DNA-VIDEO automatically performs the search for significant conformational and physico-chemical properties and the regions of sites where the mean value of a conformational and physico-chemical property in question differs significantly from the value characteristic of random nucleotide sequences.
The system B-DNA-VIDEO employs the following algorithm. Consider a nucleotide sequence S={s1…si…sL}of length L. There is the dinucleotide sisi+1 at the i-th position. The mean of the k-th property from Table 2, Xk, averaged for a region [a, b] (1<= a<= b<= L) with the starting position Ğağ and the terminal position Ğbğ of the sequence S is calculated as follows:
| (1) |
Equation (1) was applied to each of the 27 properties and to each of the 33 sites listed in Table 2. All the possible sequence regions [a, b] not smaller than one dinucleotide were taken into account. Their number for a sequence L long was n(L)=L x (L-1)/2. The total number of sequence regions was n(120)=120 x (120-1)/2=7140 for a sample. A number of properties Xk,a,b can be calculated N(L)=27 x n(L) for a sequence L long. Thus, a total number of N(70)=27 x 7140=192780 properties was tested for each sample.
Applying Equation (1) to the set of site sequences {S} at a fixed k, a and b, we yield the distribution Xk,a,b{S} for the site. Similarly, the distribution Xk,a,b{R} is generated for random sequences {R} with the same nucleotide frequencies as in the real sequences. The difference between these distributions Xk,a,b{S} and Xk,a,b{R} is tested for significance using four statistical criteria [13]. B-DNA-VIDEO [11] processes the sets {S} and {R} and outputs the list of the significant B-DNA properties {Xk,a,b}.
The results obtained by the described analysis of transcription factor binding sites are stored in the knowledge base on functional sites B-DNA-FEATURES, which is a constituent of the computer system B-DNA-VIDEO.
We have analyzed 33 transcription factor binding sites; the results are summarized in Table 2. The conformational and physico-chemical properties, the mean value of which within certain regions of a site differs significantly from the values characteristic of random sequences, have been discovered for each site considered. If the mean value of a significant property for the site exceeded that for random sequences, it was indicates by “+” in Table 2; otherwise, “-“. The lengths of the significant regions vary in the range of 10 to 25 bp; this corresponds to 1-2.5 coils of B-DNA helix as well as to the mean length of the region of DNA interaction with transcription factors (see Table 1).
An approach to classification of transcription factors based on the revealed sets of significant conformational and physico-chemical properties of their binding sites is suggested. The classification of transcription factor DNA-binding domains proposed by Wingender [14] distinguishes four superclasses: I superclass containing basic domains; II superclass, zinc-coordinated DNA-binding domains; III superclass, helix-turn-helix; and IV superclass, beta-scaffold factors with minor groove contacts. Fifteen transcription factors of thirty three considered belong to various classes of the I superclass according to the type of their domain; eleven sites, to the II superclass; and seven, to the III supercalss (Table 1).
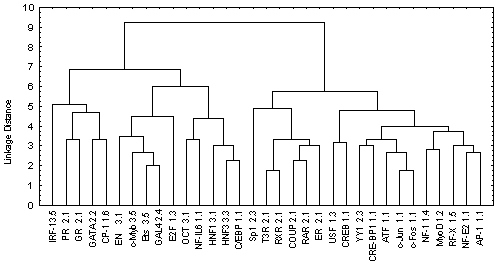
The information contained in Table 2 was processed by the program Cluster Analysis from software package STATISTICA (Windows’95) to construct the similarity tree for the transcription factors analyzed (Figure). Euclidean distance was used as a measure of similarity between two sites i and j:
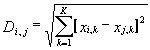 |
(2) |
where xi,k = 1, if the mean value of kth property on the region of the site i exceeds significantly the corresponding mean value for random sequences (indicated by “+” in Table 2); xi,k = -1, if the mean value of the kth property on the region of the site i is significantly lower than the corresponding mean value for random sequences (indicated by “-” in Table 2); xi,k = 0, if the mean value of the kth property on the region of site i equals the corresponding mean value for random sequences; and k is the number of the property considered, k = 1,…,27. The distances between clusters are determined by the greatest distance between any two objects in separate clusters (i.e., by the “furthest neighbors”).
Figure. The similarity tree for transcription factors.
The tree obtained has two branches (Figure). The left branch unites all transcription factors that belong to the III superclass according to the type of DNA-binding domains and eight transcription factors of the I and II superclass according to the classification of Wingender [14]. The factors belonging to the III superclass contain the DNA-binding domain of HTH type. The probability that a transcription factor is attributed to the III superclass accidentally is a <0.004. In turn, the right branch divides in two branches: one containing six transcription factors of the II superclass; the other, eleven factors of the I superclass. The probabilities of accidental attribution to these classes are ![]() <0.0014 and
<0.0014 and ![]() <0.0012, respectively. These probabilities were calculated basin of binomial distribution as follows:
<0.0012, respectively. These probabilities were calculated basin of binomial distribution as follows:
| (3) |
where m is the number of factors in the tree branch considered; n, the number of factors of a definite superclass contained in this branch; N, total number of the factors in this superclass; M, total number of factors considered (33); and P = N/M, the probability of accidental attribution of one factor to the superclass considered.
The classification of transcription factor binding sites obtained by this procedure is consistent with the X-ray and NMR structures of DNA-protein complexes as well as with other available data on DNA-protein interactions. Thus, the transcription factors of the III superclass interact with the DNA as a rule in a form of monomers forming abundant H-bonds and van der Waals (hydrophobic) contacts with bases and sugar-phosphate backbone of the DNA in both major and minor grooves as well as a plenty of water-mediated contacts. The transcription factors of the I superclass and class 2.1 (Cys4 zinc finger of nuclear receptor type) bind to the DNA in major groove in a form of homo- and heterodimers mainly through electrostatic interactions. The transcription factors belonging to the rest classes of the second superclass interact with the major groove mainly as monomers. Interaction of GATA-1 with DNA is more likely to the interaction type of HTH domains with DNA. The NMR structure of GATA-1/DNA complex indicate that the protein binds to both grooves mainly through hydrophobic interactions.
Thus, the result obtained indicates a certain correlation between the transcription binding site classification based on their significant conformational and physico-chemical properties, on the one hand, and the classification of the transcription factors binding to these sites according to their DNA-binding domains, on the other.
I am grateful to Ms. Galina Chirikova for help in translation. This work was supported by Russian Foundation for Basic Research
REFERENCES
- Suzuki M., Amano N., Kakinuma J., Tateno M. (1997) J Mol Biol., 274, 421-435
- el Hassan H.A., Calladine C.R. (1996) J. Mol. Biol., 259, 95-103.
- Suzuki M., Yagi N., Finch J.T. (1996) FEBS L., 379, 148-152.
- Shpigelman E.S., Trifonov E.N., Bolshoy A. (1993) Comput. Appl. Biosci., 9, 435-140.
- Gorin, A.A., Zhurkin, V.B., and Olson, W.K., (1995) J. Mol. Biol., 247, 34-48.
- Hogan, M.E., and Austin, R.H., (1987) Nature, 329, 263-266.
- Gotoh,O. and Tagashira,Y. (1981) Biopolymers, 20, 1043-1058.
- Satchwell S.C., Drew H.R., Travers A.A. (1986) J. Mol. Biol., 191, 659-675
- Gartenberg, M.R., and Crothers, D.M., (1988) Nature, 333, 824-829.
- Sugimoto, N., Nakano, S., Yoneyama, M., and Honda, K. (1996) Nucleic Acids Res., 24, 4501-4505.
- Ponomarenko M.P. et al., 1998, “B-DNA-VIDEO” in this issue.
- Vorobjev D.G. and Ponomarenko J.V.,1998, “SAMPLES” in this issue.
- Lehman,E.L. (1959) Testing statistical hypotheses. Willey. New York.
- Wingender E (1997) Mol Biol (Mosk), 31, 584-600.