Laboratory of Theoretical Genetics, Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 10 Lavrentiev Ave., Novosibirsk, 630090, Russia; e-mail: levitsky@bionet.nsc.ru
Keywords: bending stiffness energy, nucleosomal DNA
Nucleosome is an elementary unit of chromatin packaging and are able to play an active role in regulation of gene expression [1]. A minimal nucleosome (core) comprises eight histones: two dimers H2A-H2B and tetramer (H3-H4)2. Nucleosomal DNA wrapped around the core has a shape of solenoid containing 1.8 coils that require 145 bp. [2]. A histone H1 attached to the core-DNA complex completes the second coil on its surface. Histone H1 binds 20 additional base pairs and contributes to further chromatin compactization. Thus, a nucleosome binds about 165 bp in a more compact state of chromatin packaging. Analysis of nucleosomal DNA has so far demonstrated the existence of a weak context signal providing for precise nucleosome positioning [3-5]. According to recent X-ray structure studies, the DNA has detectable bends on the surface of nucleosome [2]. Hence, the ability of DNA to adopt certain conformation can enhance considerably its binding to nucleosome. That is why it is especially interesting to find the DNA conformational properties that are significant for nucleosome formation.
The fine structure of the profiles of bending stiffness energy of nucleosomal DNA was studied in this work. The approach based on employment of DNA persistent lengths for dinucleotide pairs [6] was used. The persistent length of DNA characterizes the length within which the DNA axis remains detectably constant. In case of dinucleotides, the parameters of persistent length [6] reflect the local contribution of a dinucleotide in the persistent length of more extended DNA region. We calculated the energy of DNA bending stiffness E as described in [6]:
| (1) |
where R = 4.3 nm is the radius of curvature of nucleosomal DNA; k, Boltzmann constant; T, absolute temperature; h = 0.34 nm, the distance between two adjacent base pairs; W, the length (bp) of a region to calculate the energy; pi, the dinucleotide parameters of persistent length (Table 1) that reflect the linear dependence between the DNA persistent length and its melting temperature [6]; and i, position of a dinucleotide in the DNA region considered. Equation (1) estimates the work necessary to wrap the chain containing W-1 dinucleotide links around the nucleosome. Thus, the smaller is the value E, the more flexible is the DNA region of length W.
Equation (1) was used to analyze 143 nucleotide sequences of nucleosome binding sites, described in the database [7]. These sites are located in various regions of the eukaryotic genome: 3′, 5′, and inner regions of the protein-encoding genes; rRNA and tRNA genes; satellite and centromeric regions of chromosomes. The site nucleotide sequences 185 bp in length (with left and right flanks of 92 bp relative to the center of the site) were extracted form the EMBL database using the codes described in [7]. Selection of such sequence length, exceeding the size of nucleosomal DNA (145 bp), is substantiated by the necessity to take into consideration possible experimental errors in determining the position of the site’s center [9]. The profile of DNA bending stiffness energy E(X) was calculated by Equation (1) for each sequence analyzed at all the possible L-W+1 positions of the scanning window, where L is the length of the sequence; W, size of the window; and X, position of the center.
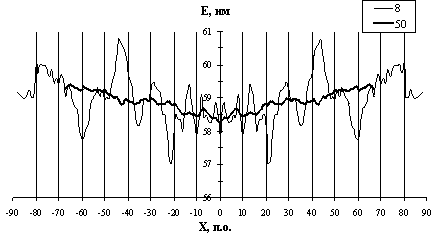
The profiles of DNA bending stiffness of all nucleosome sites [7] obtained for both orientations of nucleotide sequences were summed pointwise from 2 to 150 at window lengths being multiple of two. Thus, the symmetrized profiles E(X), mean for the sample, were determined at each length of the window (see examples in Fig. 1). Note that the profiles contain maximums and minimums, where the value E(X) differs considerably from the means averaged over the profile. Note also the dependence of the position and markedness of the extremums on the window length. The following analysis was carried out to reveal most significant minimums and maximums of the profiles E(X). The mean value Emean and dispersion ![]() were determined for the profile-forming parameters {Å(Õ)} at each window size. These parameters were used to construct the approximation of {Å(Õ)} distribution to the normal distribution N(Emean,
were determined for the profile-forming parameters {Å(Õ)} at each window size. These parameters were used to construct the approximation of {Å(Õ)} distribution to the normal distribution N(Emean,![]() ). The test on normality of distributions was positive in 72 of 75 window sizes considered; thus, the energy distribution along the profiles were considered normal. Minimum Emin and maximum Emax within the profile E(X) were determined at each window size as well as the corresponding positions of the window on the DNA. Then the distribution N(Emean,
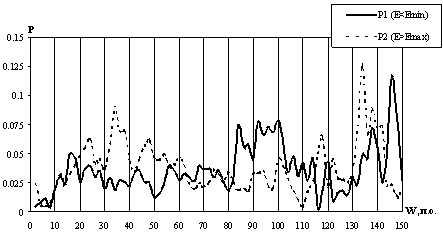
). The test on normality of distributions was positive in 72 of 75 window sizes considered; thus, the energy distribution along the profiles were considered normal. Minimum Emin and maximum Emax within the profile E(X) were determined at each window size as well as the corresponding positions of the window on the DNA. Then the distribution N(Emean, ![]() ) was used to estimate the probabilities Ð1(Å<Emin) and Ð2(Å>Emax) to obtain accidentally the energy E that is lower or higher than the minimal value Emin or maximal value Emax observed for this profile, respectively, were estimated. Depencences of the probabilities P1 and P2 on the window length are shown in Fig. 2. Eight local minimums for each of the probabilities P1 and P2, corresponding to different lengths of the window W, were revealed.
) was used to estimate the probabilities Ð1(Å<Emin) and Ð2(Å>Emax) to obtain accidentally the energy E that is lower or higher than the minimal value Emin or maximal value Emax observed for this profile, respectively, were estimated. Depencences of the probabilities P1 and P2 on the window length are shown in Fig. 2. Eight local minimums for each of the probabilities P1 and P2, corresponding to different lengths of the window W, were revealed.
It is evident from Fig 2. that the probability P1 is minimal for the windows 8, 30, 34, 40, 50, 80, 116, and 122 bp long, the lowest minimums being for the windows of 8 and 166 bp long. The position of the center of nucleosome site was taken as the origin. The centers of the regions with an increased flexibility at a 8-bp window are located at positions -21 and +21 (Fig. 1). Calculations demonstrate that these regions [-25; -17] and [+17; +25] are most flexible in the nucleosomal DNA (Fig. 1). The regions with increased flexibility revealed at the window sizes of 30, 34, 40, and 50 bp are located within one region [-26; +26]. Note that the regions revealed at a 8-bp window correspond to the ends of the region [-26; +26]. In turn, the latter region is a constituent of the regions with increased flexibility that are revealed at a longer window. Thus, the hierarchy of the regions of different lengths with increased DNA flexibility is observed within a nucleosome site.
As for the probability P2, it is minimal at window lengths of 6, 66, 70, 84, 88, 96, 110, and 148 bp. (Fig. 2). The least probabilities P2 were found for the windows of 6 and 110 bp. Regions of an increased stiffness appear at a window of 6 bp; their centers are located at positions -43 and +43 (Fig. 1). According to the calculations, it is these regions [-46; +40] and [+40; +46] that are the most stiff within the nucleosome site (Fig. 1). At a window of 60 bp, two regions with an increased stiffness were found: they are located at the right and left ends of the site at positions [-92; -26] and [+26; +92]. If the window length is increased over 66 bp, two regions of the increased DNA stiffness, also located at the ends of the site, are coming closer and overlap when the window length exceeds 96 bp. Thus, the hierarchy of the regions of different lengths and locations with increased DNA stiffness is also observed in this case.
Returning to the profile constructed for the 8-bp window (Fig. 1), note the regions of increased flexibility [-56; +64] and [+56; +64] in addition to the most flexible regions [-17; -25] and [+17; +25]. Similarly, the regions of high stiffness [-80; -72] and [+72; +80] are also evident in addition to the regions of the highest stiffness [-48; -40] and [+40; +48]. Thus, we are observing that the regions with nonaccidentally high and low flexibility are located in a contrasting vicinity, which is likely to provide the necessary conditions for DNA bending and precise positioning of the nucleosome. Analysis of the profile at a window of 8 bp in length also demonstrates that the central region about 20 bp long fail to manifest extremal bending stiffness. At longer windows, note the region of DNA flexibility [-25; +25] (50-bp window) and stiffness [-92; -26] and [+26; +92] (66 bp window).
Spatial location on the surface of nucleosome of the above-described most stiff and most flexible regions revealed at 8-bp window are shown in Fig. 3. Broken line indicate the region of DNA flexibility [-25; +25] revealed at 50-bp window; solid line, the regions of DNA stiffness [-92; -26] and [+26; +92] revealed at 66-bp window.
The conclusions of our study are consistent with the data on analysis of nucleosomes by other methods. As for the central region of the site about 20 bp in length, recent studies of nucleosomal DNA by X-ray analysis [2] and multiple alignment [4] confirm that this region is not bent. Significant periodicities of the DNA nucleosomal context were revealed in [4] in other regions of the site. Discussing the shape of bending stiffness profile of nucleosomal DNA (Fig. 1; 8-bp window), note also that it fits the shapes of the profiles of nucleosomal DNA obtained using DNA geometric parameters [5]. In that work, the authors suggest that the flexibility pattern of nucleosomal DNA consists of two bent elements 50-60 bp each that are separated by a region of smaller curvature 20-30 bp long.
The analysis performed allowed us to obtain the additional information on structural peculiarities of nucleosomal DNA. The lengths and locations of the regions within the nucleosome site that correspond to the most significant minimums and maximums of DNA bending stiffness were determined, and their hierarchy was demonstrated.
Table 1. Dinucleotide parameters of DNA persistent length [6]
|
Dinucleotide pair |
ð (nm) |
|
TA |
11 |
|
AT |
41 |
|
AA/TT |
37 |
|
CA/TG |
38 |
|
AG/CT |
43 |
|
GA/TC |
84 |
|
AC/GT |
100 |
|
CG |
63 |
|
GC |
156 |
|
GG/CC |
83 |
Fig. 1. The profiles E(X) of DNA bending stiffness at window lengths of 8 and 50 bp: X, position of the window relative to the center of the site; E, in the units of persistent length.
Fig. 2. The probabilities Ð1(Å<Emin) and Ð2(E>Emax) representing the significance of extremums of profiles E(X) depending on the window length W.
Fig. 3. Spatial location of the revealed regions contrasting in DNA flexibility on the histone octamer: (1 and 2) the most stiff regions and (3) the regions with increased stiffness; (4 and 5) the most flexible regions and (6) the regions with increased flexibility.
This work was supported by grants from the Russian Foundation for Basic Research (No.97-04-49740, 97-07-90309, 96-04-50006, 98-04-49479, 98-07-90126); Russian Ministry of Science and Technologies; Russian Human Genome Project; Russian Ministry of High Education; Siberian Department of RAS (Programms for support of reseach of young scientists and Programm of Integration projects); National Institutes of Health, U.S.A. (No.5-R01-RR-04026-08)
References
- F. Thoma,. ŦNucleosome positioning (Review)ŧ Biochim. Biophys. Acta 1130, 1-19 (1992).
- K. Luger, A.W. Mader, R.K. Richmond, D.F. Sargent, and T.J. Richmond, ŦCrystal structure of the nucleosome core particle at 2.8 A resolutionŧ Nature 389, 251-260 (1997).
- S. Satchwell, H.R. Drew, and A.A. Travers, ŦSequence periodicities in chicken nucleosome core DNA.ŧ J. Mol. Biol. 191, 659-675 (1986).
- I. Ioshikhes, E.N. Trifonov, et al., ŦNucleosomal DNA sequence databaseŧ J. Mol. Biol. 262, 129-139. (1996).
- D.J. Fitzgerald, et al., ŦConserved patterns of bending in satellite and nucleosome DNAŧ J. Biol. Chem. 269, 21303-21314 (1994).
- A.V. Sivolob and S.N. Kharpunov, ŦTranslational positioning of nucleosomes on DNA: the role of sequence-dependent isotropic DNA bending stiffnessŧ J. Mol. Biol. 247, 918-931 (1995).
- I. Ioshikhes and E.N. Trifonov, ŦNucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequencesŧ Nucl. Acids. Res. 21, 4857-4859 (1993)
- The profiles of DNA bending stiffness used in the work were built using the program Craft available at http://wwwmgs.bionet.nsc.ru/Programs/CRAFT/