LIKHOSHVAI V.A.+, MATUSHKIN Yu.G.
Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 10 Lavrentiev Ave., Novosibirsk, 630090, Russia;
e-mail: likho@bionet.nsc.ru;
+Corresponding author
Keywords: mRNA, codon distribution, translation, evolution, numerical simulation
A mathematical model describing the most general properties of the protein translation process was constructed. Regular patterns of the evolutionary drift of synonymous codons depending on the location of the codons in mRNA were analyzed theoretically. Shielding of the codons was demonstrated to impose certain selective limitations on synonymous codon usage. In particular, it follows from the model that in intensely translated mRNAs, a higher fraction of “rare” codons must be observed at the beginning of the reading frame compared to the regions more distant from the initiation codon
It is well known that the degeneracy of the genetic code leads to the ambiguity of the protein coding at the DNA level. The phenomenon of nonrandom synonymous codon usage is commonly known too. Preferential usage of some synonymous codons is recorded at different organism levels: while comparing groups of genes from different species [1], different groups of genes within one species [2], and while analyzing the frequencies of synonymous codons depending on the codon’s position in the reading frame [3]. Thus, the synonymous codon usage complies with certain selective limitations. Investigation of their nature is an important task of the theory of molecular evolution.
Several authors have studied the regular patterns of evolutionary codon drift [4, 5]. It has been demonstrated that the nonequilibrium usage of synonymous codons can be explained qualitatively basing on the general principles of the translation machinery structure and a supposition that optimization of its internal parameters aided the organism’s fitness. However, the models studied so far failed to consider the mutual arrangement of codons within the mRNA, excluding the possibility to investigate the effect of this factor on the evolution of synonymous codons.
In this work, we are constructing a mathematical model of protein translation that takes into account the steric size of ribosomes. This allows us to analyze the limitations imposed on the codon distribution along the mRNA. Results of numerical analysis suggest certain conclusions on possible mechanisms regulating the codon usage frequencies depending on their position in the reading frame. In particular, we have demonstrated that the mRNAs, the elongation rate of which tends to grow on the average from the beginning to the end on the reading frame, are evolutionarily selected; moreover, this pattern is more pronounced in intensely translated mRNAs. A relatively high frequency of relatively “slowly read” codons is, as a rule, maintained in a small region comparable in size with the steric size of the ribosome located at the beginning of the reading frame. This conclusion is in complete accordance with the Bulmer’s data on frequency analysis [3].
1. Description of the mathematical model
The processes of initiation, elongation, and translation termination in this mathematical model are described in terms of mono- and bimolecular reactions: at a formal level, the model is a system of conventional differential equations. Since the model is destined to study the regular patterns of the codon drift stemming from the most general characteristics of the translation machinery structure, no specific peculiarities of translation of individual proteins, formed in actual organisms in the course of evolution, are described. The dynamic variables of the model are the intracellular concentrations of mRNA, ribosomes, tRNA, and their complexes. Codon compositions of all reading frames, elongation constants of the codons presented to ribosomal A-sites, translation initiation constants of individual reading frames, translation termination constants, and steric size of the ribosome (which has the meaning of minimal number of codons that can share A-sites of the ribosomes translating the same mRNA) are the parameters of this model. Concentrations of ribosomes, tRNA, and individual mRNAs are the initial values specified. Numerical methods were used to calculate equilibrium values of all the dynamic variables of the model and synthesis rates of individual proteins depending on the initial values of variables and values of the parameters. The results of the computations for different values of variables and parameters were used to analyze the parametric behavior of the model of the translation system.
2. Selection of the target functional
However, it is not sufficient to construct the mathematical model of protein synthesis for solving the problem in question. It is also necessary to specify the evolutionarily meaningful criteria to estimate the parametric state of the model. This requires to assume additional hypotheses to select the target functional.
A number of natural functionals (Table) can be used to estimate various integral characteristics of the translation system. In this work, we selected the functional Vr as a target. The increase in its value caused by a synonymous mutation is interpreted in this work as an event inclined to be evolutionary fixed in the population. The model calculations have demonstrated that the parametric properties of all the functionals listed in Table are qualitatively similar enough; hence, evolutionary analysis of any functional gives qualitatively similar results. However, they can differ in some quantitative details.
Table
| N | Designation |
Brief characteristics |
Note |
| 1 | E | Index of economical efficiency | Allows the functional efficiency of the translation system to be estimated |
| 2 | Vr | Instantaneous rate of the synthesis of a protein group specified | In this work, we included in the specified group the proteins synthesized by all the reading frames. |
| 3 | Va | Instantaneous rate of amino acid residue integration in the growing peptides | |
| 4 | T | The time required to produce a fixed number of molecules of the proteins specified | Can be an estimation of the cell cycle duration for protozoan organisms |
| 5 | C | The averaged constant of instantaneous elongation rate of one codon | Can be considered for the mRNA groups or positions specified and fixed codons |
3. Results of computation and their discussion
Numerical investigation of the parametric properties of functional Vr (typical calculations are shown in Fig. 1; see the caption for calculation conditions) allows us to conclude that (1) the drift of synonymous codons is an inherent property of the translation system, that is, the codons for which the change in elongation rate is an evolutionarily significant event exist for any set of the system parameters (including the codon composition of the mRNA) and (2) a concrete parametric state of the translation system specifies unambiguously the evolutionarily significant direction of elongation rate changing (the direction of Vr increase) for a given codon, the increase in the codon elongation rate being not necessarily evolutionarily advantageous.
The numerical calculations performed have demonstrated that the instantaneous direction of the evolutionary drift of a given codon results from a complex interplay of all the parameters of the translation system. However, the following property is of the highest importance for understanding the evolutionary trends of codon drift: the increase in elongation rate of individual codons at a relatively low mRNA translation efficiency is an evolutionarily advantageous event independently of their location in the reading frame; however, the increase in the translation initiation rate above a certain threshold value (other conditions being equal) results in the situation when the decrease in the codon elongation rate in the region located at the beginning of the reading frame becomes evolutionarily advantageous; moreover, the higher is the translation efficiency, the more pronounced is this tendency-the region of relatively slow codons expands and the evolutionary benefit from the codon elongation rate decrease in these region increases (from the elongation rate of the codons located in this region).
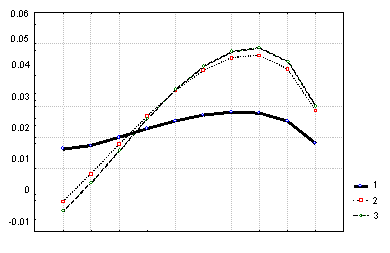
Figure 1. Changes in the protein synthesis rate with the changes in the codon elongation rate depending on the position in mRNA: in the model employed for computations, the elementary translation region is equal to the steric size of the ribosome and amounts to one codon. Abscissa, number of a conditional codon; ordinate, partial derivative of Vr with respect to the codon elongation rate. Elongation rates of all the codons are chosen equal to 1 codon/s; concentration of ribosomes, 10,000 ribosomes/cell; the fraction of the mRNA varied, 0.1% of the total pool of the translated cellular mRNA. Curve 1 shows the translation initiation constant kb=0.5 s-1; curve 2 ,
kb=1 s-1; and curve 3, kb=2 s-1.
To study the global patterns of the codon evolutionary drift, we carried out the numerical simulation of the evolutionary process. Typical results of numerical simulation (for simulation conditions, see the caption) are shown in Fig. 2. The general conclusion is the following: independently of the initial distribution of the codon elongation rates along the mRNA, the changes are evolutionary directed to gradual (on the evolutionary time scale) leveling off the elongation rates of the neighboring mRNA regions with simultaneous forming of individual elongation rate profiles, the peculiarities of these profiles being dependent on the relation of individual mRNA translation parameters and the parameters of the translation system on the whole. In particular, if the translation initiation rate of a given mRNA is high enough, the codon elongation rate remains relatively low in the region of translation initiation and then is growing along the reading frame with the distance from its beginning. A relative decrease in the codon elongation rate can be observed by the end of the reading frame. Note that in the model calculations, we observed a lower evolutionary rate of the changes in translation termination constant compared with the evolutionary rate of the changes in codon elongation constant. That is, the model predicts that the maximal speed of the ribosome movement along intensely translated mRNAs in evolutionary advanced systems is realized for the inner codons of the reading frame.
Figure 2. Changes in the codon elongation constants resulting from the evolutionary drift of the translation system toward the increase in the Vr : the calculation conditions are identical to those described in Fig. 1. The increment D =0.02 s-1 is taken as an evolutionarily fixed change of the elongation constant of the fixed codon. Abscissa, number of the codon; ordinate, the codon elongation rate constant. Curve 1, initial distribution of elongation rates with respect to the codons; curve 2, the elongation rates after 30 fixations of the changes; curve 3, after 70 fixations; and curve 4, after 200 fixations.
Thus, the most general principles of translation machinery organization together with the hypothesis on the constantly acting background factor of weak positive selection, which correlates directly with one of the natural characteristics of the translation system (in this work, this characteristics is Vr ; however, choosing of any other target functionals does not affect fundamentally the conclusion), leads to the conclusion that in the course of the evolution of an organism, the selection by the frequencies of synonymous codon usage with respect to their location along the reading frames is bound to take place. According to the general regularities of selection, the codon elongation rate tends to grow with the distance from the translation initiation site for highly expressed mRNA types. The terminal regions of the reading frame can be the exclusions. The selection fixing the codons with a low elongation rate is possible at the beginning of the gene under certain conditions. At the end of the gene, the fraction of relatively “slow” codons can also be increased; however, it is achieved due to a lower rate of “fast” codon fixation compared with the central gene region. In particular, the calculations performed have demonstrated that the selection of the translation system variants in which the translation termination rate is considerably lower than the elongation rate of an individual codon is evolutionarily advantageous. The evolutionary benefit from the increase in codon elongation rate for weakly expressed genes is relatively low and virtually independent of the codon’s position in the reading frame; hence, the effect of the steric size of ribosomes on the evolution of elongation rates along the reading frame is minimal. That is, an increase in the usage frequency of “rare” codons is bound to be observed in low expressed genes.
Note in conclusion that during the simulation of the evolution of the codon composition of translated mRNAs, we have noticed that the parametric shift of the translation system along the surface of the target functional is accompanied by leveling-off of the increments of this functional in the parameters varied. The analysis allowed us to generalize this observation as follows: the equality of the probabilities of the change in the system parameters and relative stability (absence of dramatic changes) of the environment provided, the system will evolve in such a way that the contribution of different system parameters to the increase in fitness will be approximately equal, that is, the system, in a way, “avoids” the states in which any of the parameters contribute considerably greater to the fitness than the others.
References
- S. Aota, T. Goiobori, F. Ishibashi, T. Maruyama, and T.I Kemura, “Codon usage tabulated from the Genbank Genetic Sequence Data” Nucleic Acids Res. 16 Supl., 315 (1988)
- P.M. Sharp, E. Cowe, D.G. Higgins, D.S. Shields, K.H. Wolfe, and F. Wright, “Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Scizosaccharomyces prombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity” Nucleic Acids Res. 16, 8207 (1988)
- M. Bulmer, “Codon usage and intragenic position” J. Theor. Biol. 133, 67 (1988)
- M. Bulmer, “Coevolution of codon usage and transfer RNA abundance” Nature 325, 728 (1987)
- V.A. Likhoshvai, “Rare codons: fortunity or regularity?” in “Modeling and computer methods in molecular biology and genetics” (ed. by V.A. Ratner and N.A. Kolchanov, Nova Science Publishers, Inc., USA, 1992), 463