The Sanger Centre, Hinxton, Cambridge, CB10 1SA, United Kingdom
+Corresponding author
Keywords: database, gene structures, predicted genes, large scale genome sequencing
Large scale genome sequencing projects currently produce hundreds of megabases each year. The major sequencing centers are in the process of scaling up their throughput over the next few years Shifting efforts toward sequencing gene-rich rather than random regions might provide the sequence of most of human genes during the next 3 years. Moreover, the initiative to create by 2001 a ‘rough draft’ of the human genome can allow other scientists to proceed more rapidly with discovering diseases genes. However, the sequence itself does not always provide the knowledge of gene coding regions, which are usually cover a pretty small fraction of genomic DNA. Also, we can not expect their rapid identification in near future by pure experimental approaches for such enormous volume of sequence data. The value of sequence information for biomedical community will strongly depend on availability of candidate genes computationally predicted in these sequences.
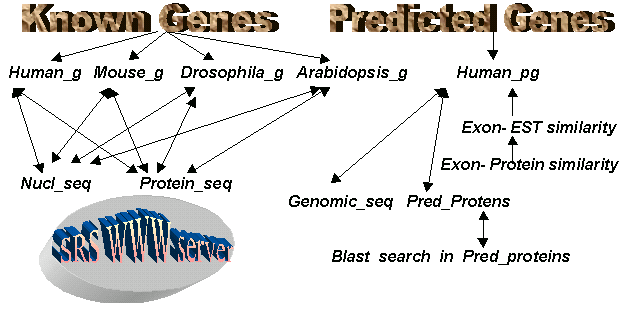
The aim of this work was to create the information resource of known and predicted gene structures in major model organisms as Human, Mouse, Drosophila and Arabidopsis. The general scheme of the INFOGENE database is presented in Fig.1.
Figure 1. General scheme of Infogene component.
INFOGENE is realized under the Sequence Retrieval System (SRS) developed in European Bioinformatics Institute (Etzold et al., 1996). This system provides a possibility to connect the database with the existing data resources (as TRRD, Transfac, Swissprot, GeneBank, etc.) and to make complex queries over several databases using WWW server. In SRS any retrieval command, logical operations with sets that were obtained by previous queries, links between sets of different databanks, or a combination of all can be easily expressed by the SRS query language.
Known gene structures Database
Primary reasons for generating known gene structure databases are:
- To have collection of known gene structures with their main features presented in the form convenient for retrieval entries including some particular features
- Easily create subset of genes or exons with a given set of features
- Check availability of genes with particular features
- Have links to different informational databases providing regulatory site locations or other information for a particular gene (about polymorphism or mutations underlying inherited disease, for example).
- Possibility to make link between similar genes of different model organisms
Today the problem of reliable gene prediction in human genomic DNA is still open. The best multiple gene prediction programs like GeneScan (Burge and Karlin,1997) and Fgenes, (Solovyev, 1998) were tested mostly on short sequences containing one gene. The recent test of these programs for 660 human genes shows that the programs can correctly predict about 80% of internal exons and just about 60% of 5’-exons (Solovyev,1988). The prediction of multiple genes should be even less accurate. Therefore, it is important for developing the further gene prediction programs to have as much as possible information about the known genes and their functional signals, that will provide the learning and testing datasets.
We have developed a GenBank parser GeneParse which produces a flat file with some description of genes and gene features including terms corresponding to exon types, regulatory elements, processes and characteristics of genes in a given GenBank sequence. To add this information to SRS we created several files with logical stricture of INFOGENE database components and files with the syntax of their entries. Using these files the information about gene structure was written to SRS with indexing of specific words in entries.
We can use the query language and search/retrieving software of SRS that will quickly extract sets of sequences with particular biological features. For example, genes where transcription start and stop sites are known or entries with multiple genes. The query language will provide an effective usage of database information in investigation of significant characteristics of genes and their regulatory elements and assist in development methods of their recognition. Currently it might take months to collect such information from the literature.
One example of INFOGENE entry corresponding MMTNFAB locus of GenBank is presented in Fig. 2. We can see that this locus includes 2 neighbor genes, which exons and coding regions were described as well as the locations of start, TATA-box and stop of transcription. In the LFT (Locus Features) field we have description of this sequence by special keywords: mang (locus includes many genes), nasp (no alternative splicing), nmts (no multiple starts of transcription) , natp (no alternative promoters), yftr (yes full transcript), npse (no pseudogenes).
For example, using ytss keyword we can easily observe that start of transcription is provided for 251 genes with completely sequenced coding regions.
LID MMTNFAB GenBank MOUSE_G DAT 19980713 LCO 7208 bp DNA ROD 11-MAY-1993 LDE Mouse complete TNF locus (TNF=tumor necrosis factor). ORG Mus musculus LKE B1 repetitive sequence; lymphotoxin; tumor necrosis factor. LFT mang nasp ytss nmts natp yftr npse LGN 2 2 2 7208 0 GID GMM000399 direct GPR TNF-beta IND PSD SWISS-PROT: P09225 GFT nasp natp ytss nmts yftr nex5 mexo fcds rsiz nsto GEC 4 3 0 0 916 609 202 609 TSS 1193 3207 1 be TAT 1174 yTAT c tata a dba EXO 1193 1345 f c gt EXO 1709 1813 i ag gt EXO 1897 1996 i ag gt EXO 2221 3207 l ag c CDS 1718 1813 f atg gt I CDS 1897 1996 i ag gt V CDS 2221 2633 l ag tag POA 3186 c aataaa c com SEQ MMTNFAB GMM000399 GRE 0 nrep dba DWG GMM000400 GUS 1669 ctccgctacacacacacactctctctctctctctcagcaggttctccaca GDS 2633 gattctaaagaaacccaagaattggattccaggcctccatcctgaccgtt GID GMM000400 direct GPR tnf-alpha IND PSD SWISS-PROT: P06804 GFT nasp natp ytss nmts yftr nnex mexo fcds rsiz nsto GEC 4 4 0 0 1691 708 235 708 TSS 4371 6968 1 be TAT 4331 yTAT c tata a dba EXO 4371 4712 f c gt EXO 5225 5279 i ag gt EXO 5457 5504 i ag gt EXO 5799 6972 l ag > CDS 4527 4712 f atg gt I CDS 5225 5279 i ag gt V CDS 5457 5504 i ag gt I CDS 5799 6217 l ag tga POA 6967 a aataaa g dba SEQ MMTNFAB GMM000400 GRE 0 nrep dba UPG GMM000399 GUS 4478 ctttcactcactggcccaaggcgccacatctccctccagaaaagacacca GDS 6217 aagggaatgggtgttcatccattctctacccagcccccactctgacccct
Figure 2. An example of INFOGENE entry corresponding MMTNFAB GenBank locus.
Database of predicted genes
Primary reason for generating predicted gene structures database is:
- Provide positional cloners, gene hunters and others with the gene candidates observed in finished and unfinished genomic sequences.
We use 2 programs GeneScan (Burge and Karlin,1997) and Fgenes (Solovyev,1998) to predict genes, because exons predicted by both programs is much more often correspond to real ones. The Blast (Altschul et al.,1977) search is used to check if some of predicted exons have similarity with known EST and protein sequences. Example of description of predicted genes is presented in Fig.3. We can see that Genescan predicted 5 coding exons (3 correct) and Fgenes predicted 5 exons (4 correct) and 1 partially correct. All exons predicted by both programs are correct.
LID HSCPH70 GenBank TEST DAT Mon Jul 13 11:44:57 BST 1998 LCO 6711 bp 0 ORG Homo sapiens 22 LKE repeats genes protein EST LFT oneg ntss mexn LGG 1 1 1 1 LGF 1 1 1 1 GID GHS000099 GFT genescan direct mexn ntss fcds GEC 5 5767bp 201aa CDS 584 632 atg gt CDS 1625 1783 ag gt HOP pir|F14571|SSC5D12 SSC5D12 NID: g972046 - pig. 6e-07 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 3e-53 CDS 4318 4406 ag gt both HOP pir|P05092|CYPH_HUMAN PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A 1e-12 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 3e-53 CDS 4628 4800 ag gt both HOP pir|P10111|CYPH_RAT PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A 2e-29 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 2e-94 CDS 6215 6350 ag tga both HOP pir|P05092|CYPH_HUMAN PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A 2e-19 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 2e-72 POA 6538 a aataaa a SEQ HSCPH70 GHS000099 GUS 584 ggcgtctctctaagatgcccaggctggtggccggtgtcgaactcctaaga GDS 6350 gtttgacttgtgttttatcttaaccaccagatcattccttctgtagctca GFT fgenes direct mexn ytss fcds GEC 5 5111bp 305aa TSS 1615 TAT 1585 yTATA CDS 1240 1728 atg gt HOP pir|F14571|SSC5D12 SSC5D12 NID: g972046 - pig 6e-04 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 5e-58 CDS 4173 4203 ag gt HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 1e-10 CDS 4318 4406 ag gt both HOP pir|P05092|CYPH_HUMAN PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A 1e-12 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 3e-53 CDS 4628 4800 ag gt both HOP pir|P10111|CYPH_RAT PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A 2e-29 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 2e-94 CDS 6215 6350 ag tga both HOP pir|P05092|CYPH_HUMAN PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A 2e-19 HOE gnl|UG|Hs#S552867 Human cyclophilin gene for cyclophilin 2e-72 SEQ HSCPH70 GHS000099 GUS 1240 aacggtcggaaggggcgtctctctaagatgctggctaattaccaggtaac GDS 6350 gtttgacttgtgttttatcttaaccaccagatcattccttctgtagctca LRE 5028 5314 AluSx SINE/Alu
Figure 3. Example of predicted gene description using HSCPH70 sequence.
The Infogene database is available through WWW server of Computational Genomics Group at http://genomic.sanger.ac.uk/