NEVINSKY G.A.+, VINOGRADOVA N.L., BUGREEV D.V., ISHENKO A.A., VASYUTINA E.L., ULYANOVA N.P., ZAKHAROVA O.D., KOLOCHEVA T.A.
Institute of Bioorganic Chemistry, Siberian Brunch of Russian Academy of Sciences, Novosibirsk, 630090, Russia
+Correspoding author
Keywords: sequence dependent and independent enzymes, the basic principles, DNA recognition mechanisms
Abstract
New methods which allow to estimate a relative efficiency of interaction of DNA-dependent enzyme with each individual nucleotide unit (and its structural elements) of any specific and nonspecific ss and ds lengthy DNA were developed. A relative contribution of internucleoside phosphate groups, specific and nonspecific bases, single chains, and the definite sequences of ds DNAs to their affinity for several repair, replication, topoisomerization and restriction enzymes was estimated. All analyzed enzymes were shown to form weak additive nonspecific contacts with internucleoside phosphate groups (and some of them hydrophobic and/ or wan der Walls contacts with the bases) of all nucleotide units of ss and/or ds d(pN)n, which are within their DNA binding clefts (depending the enzyme n = 7-20). For the 7-20-links or base pairs of long ds DNA the contribution of weak (and sometimes strong) nonspecific contacts to the specific substrate affinity was estimated to be about 4-7 orders of magnitude and predominate over the specific ones (0.5-2 orders of magnitude) due to additivity of free energies of individual nucleotide units recognition.
As was shown, the complex formation of sequence-dependent enzymes with cognate DNAs can not provide specificity of the enzyme’s action. The high specificity of the enzyme functioning may be provided by the DNA adaptation processes and the kinetic constant of the reaction. Transition from nonspecific to specific DNA leads to the increase of the kinetic constant by 4-7 orders of magnitude. The enhancement of DNA substrate rate conversation occurs if molecule of DNA can change its structure due to interaction with enzyme to the optimal for the catalysis.
Introduction
Many sequence dependent (S-D) enzymes have been shown to be involved in various biological processes such as a DNA replication, transcription, repair, recombination, and chromosome dynamics, and are the pharmacological targets of a number of antiviral and antitumor drugs. Therefore a better understanding of the molecular mechanisms of their functioning is important from fundamental and applied points of view. During the past years, significant progress has been achieved in the study of nucleic acid interactions with the site-specific proteins owing to the development of genetical, biochemical, and different physico-chemical methods including X-ray analysis, which is known to be the most informative method. It should be noted, that detailed analysis of X-ray data of many S-D enzymes led to conclusion that specific interactions between the proteins and DNA (like pseudo-Watson-Crick interactions) are most important for specific recognition of strictly defined DNA sequences and that such contacts can provide as high affinity so as specificity of enzyme’s action ([1] and refs. therein). However, such concepts steam from X-ray data lack a sound quantitative basis. Unfortunately, there is practically no quantitative data on the contribution of the thermodynamic (complex formation) and kinetic (rate constant) factors of the catalytic process to the affinity and specificity of sequence-dependent enzymes. We failed to find any quantitative data on a relative contribution of specific and nonspecific interactions in DNA recognition by S-D enzymes. At the same time, in our opinion only detailed quantitative analysis of a relative contribution of all contacts and of all most important factors (complex formation, adaptive conformational changes in DNA, and catalysis) may help to correctly interpret the qualitative X-ray patterns of DNA-protein contacts.
Results and discussion
For analysis of nucleic-acid interactions we have developed during last years the new approaches which allow to estimate a relative contribution of each nucleotide unit of DNA to its total affinity for analyzed enzyme ([1-7] and refs. therein). We have shown that fine mechanisms of protein-DNA interactions at the molecular level can be analyzed using methods of synthesis and analysis, i.e. the slow, step-by-step complication or simplification of the analyzed ligand structure according to the scheme: dNMP or orthophosphate as the minimal ligands![]() ss homo-d(pN)n
ss homo-d(pN)n![]() ss hetero-d(pN)n
ss hetero-d(pN)n ![]() ss specific hetero-d(pN)n
ss specific hetero-d(pN)n![]() ds homo-d(pN)n
ds homo-d(pN)n![]() ds hetero-d(pN)n
ds hetero-d(pN)n![]() ds specific hetero-d(pN)n
ds specific hetero-d(pN)n![]() specific long DNA.
specific long DNA.
The additivity of enzymes interaction with various structural elements was shown to hold practically on all steps of the complication for all diapason of the ligands used.
The recognition of DNA by various S-D repair [uracil- (UDG), 8-oxoguanosine- (8-oG-DG), and hypoxanthine DNA glycosylase; apurine-apyrimidine endonuclease (AP-EN)], topoisomerization [human and mouse DNA topoisomerase I (Topo I)], and EcoRI restriction endonuclease, as well as sequence-independent pro-, eucaryotic, viral and archaebacterial DNA polymerases, and human DNA ligase have been investigated using the new approaches. It was shown that all these enzymes interact with all mononucleotide units of ss and/or ds DNA, which are within the DNA-binding clefts of the enzymes. The DNA-binding sites of the enzymes consist of the 7-20 (n) subsites interacting with n nomonucleotide units or base pairs of ss and ds DNA: n = 9-12 in case of all repair enzymes and Topo I; 7-8 for EcoRI, and 20 or 10 for templates and primers of DNA polymerases, respectively. One of the 7-20 subsites (usually active center or additional specific site) of all enzymes interacts with dNMP or orthophosphate demonstrating a relatively high affinity to such minimal ligands. In all cases transition from the minimal ligands to ss d(pN)n and then to ds DNA, the affinity gradually rises so that the dependence of lgKd upon the number of nomonucleotide unit (n) is linear up to n <= 7-20 (as an example see Figure), showing the additive contribution of all d(pN)n links to the DNA-ligand affinity. The Kd values, characterizing interaction of any enzyme with ss and ds d(pN)n, change according to the following decreasing geometric progression:
Kd[d(pN)n] = Kd[(Pi)]· [1/f]n = Kd[(dNMP)]· [1/f]n-1 (I)
where Kd(1) and Kd[d(pN)n] – Kd values for the minimal ligand of enzyme and for d(pN)n, respectively; f = 1/Kd is the factor of d(pN)n affinity increase (it changes from 1.3 to 2.7 with different enzymes and other factors, see below) due to lengthening of ss or ds d(pN)n by one nucleotide unit or by one base pair. (7 <= n <= 20)
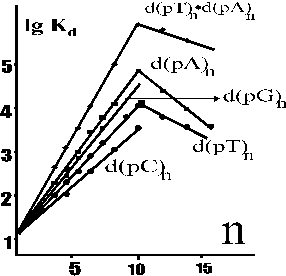 |
Figure. The logarithmic dependencies of the Kd values of Topo I complexes with ss and ds d(pN)n upon the number of the links (n). |
The f-values can be calculated from the slopes of the dependencies of lgKd vs n for ss and ds d(pN)n-ligands. As was shown by different methods, the increase of the DNA affinity for 8-oxoG-DG, AP-EN and EcoRI takes place due to formation of weak additive electrostatic contacts between the enzymes and internucleoside phosphate groups of DNA. These enzymes do not interact with the bases of ss and ds DNA. The slopes of the logarithmic dependencies of the Kd values on the number of nucleotide units (n) for 8-oxoG-DG, AP-EN and EcoRI are the same for all homo-, hetero-d(pN)n, and d(pR)n oligomers without bases (R is chemically stable analog of deoxyribose, where 1’-base was displaced for hydrogen atom).
All DNA polymerases, Topo I and UDG form also weak nonspecific additive electrostatic contacts with internucleoside phosphate groups of ss and ds DNA. But, in contract to the above mentioned enzymes, a relative efficiency of their interaction with DNA correlate with a relative
hydrophobicity of the DNA nucleotide bases (C< T< G< A): the enzymes form hydrophobic and/or van der Waals contacts with bases of DNA molecules. It should be stressed that the interaction of all enzymes analyzed with ss DNA may be described by common algorithm:
Kd[d(pN)n]= Kd[(Pi)]· [f]-n = Kd[(Pi)]• (е)-n• (hC)-l• (hT)-m• (hG)k• (hA)-g (II)
where e is the electrostatic incremental factor for d(pN)n extension by one internucleoside phosphate group; hC, hT, hG, and hA – factors, reflecting increase of the affinity due to hydrophobic interaction of enzyme with one of the C, T, G, or A bases of ss DNA (n= l+ m+k+g). It should be noted, that the efficiency of the interaction of S-D or sequence-independent enzymes with any nonspecific DNA (even when DNA contains the most part of the specific sequence) do not practically depend on their contexts; the affinity of such DNAs depend only upon the number of C, T, G, and A bases and/or internucleoside phosphate groups into ss or ds DNAs. The numerical values of the Kd[(Pi)], f, е, and hN values are different for various enzymes and in case of ss or ds DNA: they are given in the Table.
We have analyzed the influence of the second complementary strand on the enzyme interactions with the first chain. UDG and EcoRI was shown to melt completely ds d(pN)n (n<= 15-20), and the second chain did not increase the affinity of the first one (Table).
Table. Kinetic and thermodynamic data on various enzymes interaction with DNA
|
Enzyme |
||||||
| Ligand or parameter | 8-oG-DG | EcoR1 | UDG | Topo I | Klen. Fr.,Template | Klen. Fr.,Primer |
| Pi, Kd (M) | 1.0· 10-2 | 1.5· 10-2 | 1.7· 10-2 | 7.0· 10-2 | 2.6· 10-5 | 7.0· 10-5 |
| dTMP, Kd (M) | 8.0· 10-2 | 1.0· 10-3 | 1.5· 10-2 | 5.0· 10-2 | 1.0· 10-5 | 3.9· 10-5 |
| n (the number of subsites) | 10-13 | 7-8 | 10 | 10 | 19-20 | 9-10 |
| n-spec. Ss d(pN)n; Kd(1),(M) | 3.3· 10-5 | 2.1· 10-5 | 8.3· 10-5 | 1.4· 10-5 | 5.0· 10-10 | only |
| specific ss d(pN)n; Kd(2), (M) | 2· 10-6 | only ds | 3.0· 10-5 | 9.0· 10-8 | 5.0· 10-10 | complem. |
| The ratio of Kd(1)/Kd(2) | 15-17 | – | 3.0-5.0 | 156.0 | 1.0 | chain |
| fC (hC, hydrophobic factor) | 1.5 (1.0) | 2.0 (1.0) | 1.43 (1.08) | 1.71 (1.06) | 1.58 (1.04) | only |
| fT (hT, hydrophobic factor) | 1.5 (1.0) | 2.0 (1.0) | 1.60 (1.22) | 1.93 (1.19) | 1.78 (1.17) | complem. |
| fG (hG, hydrophobic factor) | 1.5( 1.0) | 2.0 (1.0) | 1.77 (1.31) | 2.12 (1.31) | 1.95 (1.28) | interact. |
| fA (hA, hydrophobic factor) | 1.5 (1.0) | 2.0 (1.0) | 1.92 (1.41) | 2.30 (1.42) | 2.0 (1.32) | with |
| electrostatic factor e | 1.5 | 2.0 (1.0) | 1.35 | 1. 62 | 1.52 | template |
| n-spec. Ds d(pN)n; Kd(1),(M) | 3.0· 10-7 | 5.0· 10-6 | 9.0· 10-5 | 1.2· 10-6 | 1.0· 10-11 | 1.2· 10-9 |
| specific ds d(pN)n; Kd(2),(M) | 1.0· 10-8 | 5.0· 10-8 | 1.0· 10-5 | 5.0· 10-9 | 1.0· 10-11 | > 105 |
| the ratio of Kd(1)/Kd(2) | 30.0 | 5-100 | 3-10 | 240.0 | 1.0 | – |
| the ratio of [kcat (specific)/kcat (nonspecific)] |
105-106 | 105-106 | 5-50· 103 | 105-106 | – | |
| fHB , icrease of affinity dueto 1 HB with second strand | 1.27 | 1.0 | 1.0 | 1.13 | 1.20 | 1.35 |
Topo I,8-oG-DG, AP-EN, and DNA polymerases interact with both strands of ds DNA and the strands interact to each other, but due to a partial melting of ds DNA there is more than one order of magnitude weakening of efficiency of formation of Watson-Crick hydrogen bonds between complementary bases as compared with that in solution. In all cases the contribution of the second strand to the affinity of ds DNA is significantly smaller than that for the first one. The increase of enzyme affinity for the second chain of ds DNA (or for ds DNA in comparison with ss DNA) due to its interaction with protein and complementary interactions between the chains may be described by a decreasing geometric progression:
Kd[second d(pN)n ] = Kd[minimal ligand; usually dNMP]· (fHB)1-n (III),
where fHB is a factor of second strand affinity increase doe to one hydrogen bond formation with the first chain (Table). When affinity of minimal ligand (interacting with one of the enzyme specific subsites for recognition of the second strand) is very low (Kd![]() 1 M), the enhancement of the duplex affinity (in comparison with ss DNA) takes place only due to weak additive interaction of the second chain with enzyme and weak complementary interactions with the first chain. In fact, the interaction of enzymes with first chain leads to change of its conformation to the conformation of ds DNA and the addition of the second complementary strand to a ss DNA result mainly in an increased number of contacts with the preservation of those involved in the enzyme interaction with the first strand. And the efficiency of contacts with individual structural elements of each strand within ds DNA is comparable to that for low-molecular-weight ligands incorporating the same structural elements. The interaction of ds DNA with enzymes may be described using the same algorithms (I – II ), but due to contribution of two nucleotide units of base pairs, the numerical values of f , e and fN factors may be slightly higher. Taken together, it should be stressed that the contribution of non-specific weak and strong additive electrostatic, hydrophobic and/or van der Waals contacts of enzymes with internucleoside phosphate group and/or bases of both strands of dsDNA can reach 5-7 orders of magnitude (Table).
1 M), the enhancement of the duplex affinity (in comparison with ss DNA) takes place only due to weak additive interaction of the second chain with enzyme and weak complementary interactions with the first chain. In fact, the interaction of enzymes with first chain leads to change of its conformation to the conformation of ds DNA and the addition of the second complementary strand to a ss DNA result mainly in an increased number of contacts with the preservation of those involved in the enzyme interaction with the first strand. And the efficiency of contacts with individual structural elements of each strand within ds DNA is comparable to that for low-molecular-weight ligands incorporating the same structural elements. The interaction of ds DNA with enzymes may be described using the same algorithms (I – II ), but due to contribution of two nucleotide units of base pairs, the numerical values of f , e and fN factors may be slightly higher. Taken together, it should be stressed that the contribution of non-specific weak and strong additive electrostatic, hydrophobic and/or van der Waals contacts of enzymes with internucleoside phosphate group and/or bases of both strands of dsDNA can reach 5-7 orders of magnitude (Table).
We have estimated the contribution of specific sequences and bases to the total affinity of DNA for the enzymes (Table). During enzyme-dependent change of DNA conformation, special subsites of S-D enzymes form additional specific contacts with cognate DNAs, but such enzymes keep the main part of previously generated nonspecific additive contacts. It should be emphasized, that the limit of the contribution of specific contacts to the enzymes affinity for DNA does not exceed of 1-2 orders of magnitude in comparison with 5-7 orders of magnitude coming from the nonspecific interactions. Thus, it is evidently that complex formation between the enzymes and specific DNA can not provide specificity of their functioning.
In order to reveal why in spite of such situation with complex formation S-D enzymes analyzed are very specific, we analyzed the kinetics of their functioning. It was shown that the high specificity of the enzyme functioning may be provided by the DNA adaptation processes and the kinetic constant of the reaction. The enhancement of DNA substrate rate conversation occurs if molecule of DNA can change its structure due to interaction with enzyme to the optimal for the catalysis. Transition from nonspecific to specific DNA led to increase of the kcat by a factor of 104-107 (Table). A comparison of the result of thermodynamic and kinetic study of UDG, Topo I and DNA polymerase with X-ray analysis data of these enzymes have shown that data obtained using first approach are in agreement with the results of second one. Nevertheless, the conclusions coming from crystallographic investigation and concerning of an importance of various factors for recognition and specific conversation of specific DNA substrate may be significantly corrected. Thus, the most important factors of DNA recognition by various S-D and independent enzymes were revealed and the new concept of DNA recognition was proposed.
References
- Nevinskii G. A., “Important role of weak interactions in long DNA and RNA molecule recognition by enzymes”, Mol. Biol. (Moscow) 29, 6-19 (1995).
- D.G. Knorre, O.I. Lavrik, G.A. Nevinsky, “Protein-nucleic acid interaction in the reactions catalyzed with DNA polymerases”, Biochimie 70, 655-661 (1988).
- G.A. Nevinsky, A.G. Veniaminova, A.S. Levina, V.N. Podust, O.I. Lavrik, E. Holler, “Structure-function analysis of mononucleotides and short oligonucleotides in the priming of enzymatic DNA synthesis” Biochemistry 29, 1200-1207 (1990).
- G.A. Nevinsky, M-L. Andreola, V.I. Yamkovoy, A.S. Levina, Ph.J. Barr, L. Tarrago-Litvak, S. Litvak, “Functional analysis of primers and templates in synthesis of DNA catalyzed by HIV-I immunodificiency virus type 1 reverse transcriptase”, Eur.J.Biochem, 207, 351-358 (1992).
- G.A. Nevinsky, D.V. Bugreev, V.N. Buneva, Y. Yasui, M. Nishizawa, T. Andoh “High affinty interaction of mammalian topoisomerase I with short, single and double-stranded oligonucleotides with specific sequences”, FEBS Lett. 368, 97-100 (1995).
- T.I. Kolocheva, G.A. Nevinsky, A.S. Levina, V.V. Khomov and O.I. Lavrik, “The mechanism of recognition of templates by DNA polymerases from pro- and eukaryotes as revealed by affinity modification data”, Journal of Biomolecular Structure and Dynamics 9, 169- 186 (1991).
- T.I. Kolocheva, G.A. Maksakova, O.D. Zakharova and G.A. Nevinsky, “The algorithm of estimation of the Kd values for primers in DNA synthesis catalyzed by human DNA polymerase
 “, FEBS Lett. 399, 113-116 (1996).
“, FEBS Lett. 399, 113-116 (1996).
