BUGREEV D.V.+, VASYUTINA E.L., BUNEVA V.N., NEVINSKY G.A.
Novosibirsk Institute of Bioorganic Chemistry, Siberian Division of the Russian Academy of Sciences, Novosibirsk 630090, Russia;
+Corresponding author
Keywords: DNA topoisomerase I, recognition mechanism, oligonucleotide
Abstract
Interaction of human sequence-dependent DNA topoisomerase I (topo) with specific and non-specific oligodeoxynucleotides (ODNs) of different length and structure was investigated. The most important structural elements of DNA involved in protein-DNA interaction were revealed. Topo was shown to interact with nonspecific ODNs forming weak additive electrostatic (internucleoside phosphate groups) and hydrophobic and/or van der Waals contacts (bases of DNA) with 10 nucleotides of ss and with 10 base pairs of ds DNA These non-specific contacts provide 4 and 5 orders of magnitude in affinity of topo I for ss DNA (Kd=10-4 M) and ds DNA (Kd=10-5 M), respectively. Transition from nonspecific ss d(pN)10 to specific ss d(ApApApGpApCpTpTpApG) leads to the reinforcing of the enzyme contacts practically with all nucleotides of this ODN, but interaction of topo I with its pGpA and pT parts is most important for specific recognition by the enzyme of the scissile chain. Additional interactions of the specific ss ODN with topo (10-1M) and complementary interactions with the second nonscissile chain (10-1M) increase its affinity by two orders of magnitude (Kd=10-7M). Preincubation of ss and ds specific ODNs with topo causes the change of the ligand structure and as a consequence an enhancement of their affinity by one order of magnitude (10-1M). As a result the affinity of 27-unit-long specific ds ODN (due to sum of specific and additive nonspecific interactions) reaches 10-8 M. Finally, the enzyme affinity for supercoiled (sc) DNA (Kd=10-10 M) is about two orders of magnitude higher than for relaxed one. This two orders of the affinity can be assigned to the stronger curvature of specific sequence in sc DNA than in its relaxed form. In this way, the high affinity of topo to sc DNA is the product of these four above factors most important for recognition. A correlation between the thermodynamical findings and the X-ray studies of enzyme was revealed. A possible mechanism of topo action has been proposed.
Introduction
Eukaryotic DNA topo is an abundant nuclear enzyme that controls and modifies the topological states of negatively and positively sc DNA in such key nuclear processes as DNA replication, transcription, recombination, and chromosome dynamics [1]. The pharmacological interest to this enzyme is motivated by the fact that topo is the specific targets of a number of antitumor drugs. Thus, a better understanding of the enzyme’s function and molecular mechanism is important from both fundamental and applied points of view. During past years a significant progress has been made in the study of the protein-DNA interaction by different physico-chemical methods including X-ray analysis, which is known to be the most informative method. Resent X-ray study of topo in complex with DNA has revealed many aminoacid residues of the enzyme that contact with DNA and form the catalytic center, a possible mechanism of DNA cleavage and relaxation also has been proposed [2,3]. All X-ray analysis data obtained have qualitative character and in literature there is no quantitative data on the contribution of the contacts revealed as well as the thermodynamic (complex formation) and kinetic factors as a whole to the recognition process. However, from our point of view correct interpretation of X-ray analysis may be done only on the basis of thermodynamical and kinetical study of the enzyme.
Results and discussion
To analyze the topo-DNA interaction we have developed the inhibitory analysis approach which allow to estimate a relative contribution of each nucleotide unit of DNA to its total affinity for enzyme [4]. We have shown that the fine mechanisms of protein-DNA interactions at the molecular level can be analyzed using methods of synthesis and analysis, i.e. the slow, step-by-step complication or simplification of the analyzed ligand structure according to the scheme: dNMP or orthophosphate as the minimal ligands![]() ss homo-d(pN)n
ss homo-d(pN)n ![]() ss specific hetero-d(pN)n
ss specific hetero-d(pN)n![]() ds homo-d(pN)n
ds homo-d(pN)n ![]() ds specific hetero-d(pN)n
ds specific hetero-d(pN)n![]() specific long DNA. Using our findings [4] and recent X-ray studies [2,3] the complete pattern of topo I-DNA interaction can be proposed.
specific long DNA. Using our findings [4] and recent X-ray studies [2,3] the complete pattern of topo I-DNA interaction can be proposed.
Interaction of topo with non-specific ODNs. Since cleavage in DNA occurs only at sites situated in regions with potential for intrastrand base-pairing due to distal complementary sequences, topo has been considered to be a ds-specific enzyme which can only recognized DNA of reasonable length, and it was assumed that topo cannot interact with short ssODNs. We have therefore investigated recently the possibility that mammalian topo can form specific and non-specific complexes with short ss ODNs [4]. The results demonstrate that both specific and non-specific ODNs have a high affinity for the enzyme and are efficient inhibitors of topo-dependent relaxation of sc DNA. As for replication and repair enzyme, the minimal ligand for topo is orthophosphate (Kd ![]() 0.1 M). Proceeding from Pi to dNMP and further to d(pN)n, the affinity gradually rises so that the dependence of lg(Kd) on the link number is linear to n<= 10, testifying to additive contribution of each nucleotide unit of the 10-long-length ODNs. For each type of homooligonucleotide the change in affinity is described by a decreasing geometric progression:
0.1 M). Proceeding from Pi to dNMP and further to d(pN)n, the affinity gradually rises so that the dependence of lg(Kd) on the link number is linear to n<= 10, testifying to additive contribution of each nucleotide unit of the 10-long-length ODNs. For each type of homooligonucleotide the change in affinity is described by a decreasing geometric progression:
Kd[d(pN)n] = Kd[(Pi)] [1/f]n,
where f is the incremental factor for d(pN)n extension by one link. The latter values (f) can be obtained from the slopes of the dependence of lg(Kd) upon the number of nomonucleotide unit (n). The f-factors were found to be equal to: 1.71 – d(pC)n,1.93 – d(pT)n, 2.12 – d(pG)n, 2.30 – d(pA)n, 2.63 – duplex of d(pT)n and d(pA)n. The f -values increase with a relative hydrophobicity of the nucleotide bases of ODNs: C<T<G<A. The extrapolation of the dependence of lgf values as a function of a relative hydrophobicity C, T, G, and A nucleosides to zero gives factor e =1.64 This value (e) is the electrostatic incremental factor for d(pN)n extension by one internucleoside phosphate group. The difference of f and e factors gives factor h (h = f/e) that reflects the increase of the affinity due to hydrophobic and/or van der Waals interactions of topo with the bases of DNA. By this means, topo forms contacts with the 10 phosphate groups and with the 10 bases. This interaction of topo with any nonspecific ss d(pN)n (and ds ODNs, see below) may be described by common algorithm:
Kd[d(pN)n]= Kd[(Pi)] [f]-n = Kd[(Pi)] (х)-n (hC)-l (hT)-m (hG)-k (hA)-g,
where hC, hT, hG, and hA – factors, reflecting increase of the affinity due to hydrophobic interaction of enzyme with one of the C (hC =1.04), T (hT =1.18), G (hG =1.29), or A (hA =1.40) bases of ss DNA (n=l+m+k+g). Taken together, it is obviously that a relative contribution of additive electrostatic interactions of topo with internucleoside phosphate groups of ss ODNs predominate over the additive hydrophobic interactions and only in the case of the most hydrophobic d(pA)n the relative contributions of electrostatic and hydrophobic weak interactions become to be comparable. Our data are in agreement with X-ray analysis results: the central pore of the topo molecule is composed largely of positively charged residues, 15 lysines and 8 arginines, which give rise to a high positive electrostatic potential surrounding the pore. Thus, weak additive interaction of internucleoside phosphate groups of DNA with the enzyme may be realized through the interaction between the surfaces having opposite net charges. However, in the DNA binding cleft of topo there are not only positively charged aminoacid residues. Weak hydrophobic and/or van der Waals interactions of DNA with the enzyme may be provided due to the bases interaction with both non-charged aminoacids and non-polar parts of positively charged residues.
The addition of the second complementary chain results in its interaction with the enzyme and formation of Watson-Crick hydrogen bonds with the first chain. It is obviously that positively charged surface of the enzyme pore in principle can provide weak electrostatic interactions and a high affinity to both first and second chains of ds DNA. At the same time, on the level of relatively short d(pN)n (n <= 20), which are within the enzyme binding cleft and there is no the duplex stabilization by complementary interactions out of the enzyme, the contribution of the second strand to the affinity of the first one is very small: the incremental factor of one A-T pair formation was estimated to be 1.14 (factor fHB). It is not possible to exclude that complementary interaction between the bases of the strands may result in a decrease of the hydrophobic interaction of topo Iwith the bases of the first chain. If this is the case, the factor fHB characterizing the apparent increase of the duplex affinity in comparison with ss ODN may be underestimated. Nevertheless if to take this assumption into account, a possible maximal value of fHB factor calculated by multiplying fHB (1.14) and hA (1.40) may be estimated equal to 1.6. But even in this case, the complementary interactions between the duplex strands in the complex with topo are more than one order of magnitude weaker than that in solution. Thus, the only way to explain this observation to suppose that the interaction between DNA and topo leads to a partial melting of ds DNA and to a significant weakening of the complementary interactions (on retention of the DNA B-form, revealed by the X-ray analysis). Such weakening seems to be the main reason of a small contribution of second chain to the topo affinity for the duplex. Finally, the weak additive interaction of the enzyme with both strands of DNA and weak complementary interactions between the strands provide 4 and 5 orders of the affinity of ss (Kd=10-4 M) and ds DNA (Kd =10-5 M), respectively. Preincubation of nonspecific ODNs with the enzyme leads to the additional increase of their affinity by a factor of about 3.0
Interaction of topo I with specific ODNs. The enzyme-mediated DNA cleavage requires an asymmetric region encompassing the cleavage site. The minimal region consisted of obligatory nine nucleotides on scissile strand and five nucleotides on the non-scissile strand, with the majority of the nucleotides situated upstream to the cleavage site has been established.
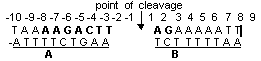
Topo-mediated cleavage also required bipartite interaction with a duplex region around the cleavage site (region A) and another duplex region on the side holding the 5’-OH end generated by cleavage (region B). According to X-ray analysis data the nucleotide units of the scissile chain can form contact with the next aminoacids of the enzymaaaaae:
In order to estimate a relative contribution of every nucleotide of the sequence to specific DNA interaction with topo, we examined how human topo recognizes different specific ODNs including dinucleotides. In contrast to nonspecific d(pC)2, d(pT)2, d(pG)2, and d(pA)2 (Kd=0.1-0.07 M) only pGpA dinucleotide (positions -5, -4) demonstrated extremely high affinity for topo (Kd=9З10-5 M). All other di- and trinucleotides corresponding to various parts of the sequence (TAG, CTT, ACT, GAC), and even tetra (GACT, ACTT) and pentanucleotides (CCTAG) were significantly worse inhibitors of the enzyme (1З10-4-1З10-3 M). This data showed that topo interacts more effectively with the pGpA dinucleotide than with other shorts ODNs corresponding to different parts of the specific sequence. Since nonspecific dinucleotides can not interact with the enzyme showing affinity comparable with that for the pGpA dinucleotide, one can suppose that interaction of the enzyme with this dinucleotide has specific cooperative feature. All data obtained speak in favor of the enzyme cooperative interaction not only with the pGpA sequence but also with other different ODNs of the sequence. Most probably, that specific contact of topo with the pGpA dinucleotide may be the initial stage of recognition initiating its following interaction with the complete specific sequence. From our point of view Pi and various dNMP as a minimal ligands of topo form contacts with aminoacid residues interacting with internucleoside phosphate of the GpA dinucleotide within the specific sequence. Interestingly, that the addition to the pGpA of two 5’-terminal A-nucleotides leads to a remarkable increase of the ODNs affinity (Kd=1.5З10-5 M, 1З10-5 M, AGA and AAGA, respectively). According to X-ray analysis data internucleoside phosphate groups corresponding to pC (-3) and pT (-2) links of the sequence form electrostatic contact with the enzyme. In spite of it, pC and pT nucleotides of GAC (Kd=1З10-4 M) and GACT (Kd=2З10-4 M) ODNs do not interact with the enzyme. In addition, these nucleotides possess negative cooperative effect on topo interaction with the pGpA dinucleotide sequence. It should be noted that according to X-ray data there is only one base-specific contact of topo with the (-1)-T base of the scissile strand. Since GACTT (Kd=2З10-6 M) pentanucleotide demonstrates about two orders of magnitude higher affinity that that for the pGpA dinucleotide. It seems reasonable to suppose that the specific contacts of this T-nucleotide are very important for effective interaction of topo with CTT-part of the sequence. There is no remarkable change of the affinity at transition from GACTT pentanucleotide (Kd=2З10-6 M) to specific AAGACTTAG nonanucleotide (Kd=1.6З10-6 M). Most probably specific contacts of the enzyme with the 3’-terminal AG nucleotides of nonanucleotide can not be enough effective on the level of short ss ODNs. Additional increase of the affinity of specific 5-9-link-ODNs occurs after addition of the complementary ODNs of the same length corresponding to non-scissile strand. Mixtures of complementary penta- and nona-ODNs effectively inhibit topo at a lower concentration than the individual ligands, although the melting points of these ds ODNs are lower than the reaction temperature. Since such short ds ODNs cannot exist in solution at 30oC, we believe that topo can combine and stabilize them as a duplex after initial step of recognition. Apparently this effect can be observed when partially complementary ODNs are used. We suggest that the second strand can increase the efficiency of scissile strand recognition by bringing optimal conformation when the phosphate contacts and specific contacts with essentially bases can be completely realized.
Long 27-unit specific duplex demonstrates the highest affinity to the enzyme. There are two possible reasons of such observation. The first one is that A-specific region of long specific duplexes can be absolutely adopt to topo recognition center. And the second one concern with specific structure of B-DNA region containing (dA) (dT)-tracts associated with helix curvature which as was found [5] to stabilize topo-DNA interaction. As was shown by us, there is no increase of the affinity of nonspecific d(pN)n at n>10. We supposed the topo binding site is like a arc form with about a straight central part and bending flanks; it can adopt only shot n<= 10 ODNs. The long nonspecific ODNs having no curved structure most probably cannot effectively interact with all parts of topo binding site, which covers 20 base pairs of ds DNA.
Adaptation of DNA to optimal conformation. The preincubation of ss and ds ODNs of different length decreased the Kd values by a factor of 3-10. This factor was about 2-3 times higher for specific ODNs than for non-specific ODNs. Most significant increase of the affinity (15-42 times) occurred when partially complementary long ODNs were used. This data speak in favor of topo-mediated transformation of the structure of the ODNs to the optimal conformation following the initial step of their recognition. More likely the adaptation should be carried over to conformation changes of cleavable phosphate group which form specific contacts with Arg488, Arg590 and His632 [3] to prepare this phosphate group for nucleophilic attack by Tyr723. The relative contributions of various parts of the sequence and other factors to the total affinity of sc DNA are shown on the above scheme.
Two orders of magnitude of overall scDNA affinity is provided by topological stress. The affinity of 27-unit-long specific ds ODN (due to sum of specific and additive nonspecific interactions) reaches 10-8 M (as for relaxed DNA). Finally, the enzyme affinity for sc DNA (Kd=10-10 M) is about two orders of magnitude higher than for relaxed one. Since dynamic curvature in sc DNA may substitute for stable curvature, this two orders of affinity can be assigned to topological stress.
References
- J. C. Wang, “DNA topoisomerases” Annu. Rev. Biochem. 65, 635-692 (1996)
- M. R. Redinbo, L. Stewart, P. Kuhn, J. J. Champoux, W. G. Hol, “Crystal structures of human topo I in covalent and noncovalent complexes with DNA” Science 279, 1504-1513 (1998)
- L. Stewart, M. R. Redinbo, X. Qiu, W. G. Hol, Champoux J. J. “A model for the mechanism of human topoisomerase I” Science 279, 1534-1541 (1998)
- D. V. Bugreev, E. L. Vasiutina, G. A. Maksakova, V. N. Buneva, T. Ando, G. A. Nevinskii, “Recognition of human DNA topoisomerase I by superhelical DNA: evaluation of the relative contribution of specific and nonspecific interactions” Mol Biol (Mosk) 31, 418-430 (1997)
- S. Krogh, U. H. Mortensen, O. Westergaard, B. J. Bonven “Eukaryotic topoisomerase I-DNA interaction is stabilized by helix curvature” Nucleic. Acids. Res. 19, 1235-1241 (1991)

