KANYSHKOVA T.G.+, SEMENOV D.V., BUNEVA V.N., NEVINSKY G.A.
Institute of Bioorganic Chemistry, Siberian Brunch of Russian Academy of Sciences, 8 Lavrentiev Ave., 630090, Novosibirsk
+Corresponding author
Keywords: transcriptional activation, human lactoferrin, protein-DNA interactions, polyanion binding
Abstract
Many different unique functions have been attributed to lactoferrin (LF), including protection from iron-induced lipid peroxidation; immunomodulation, cell growth regulation; DNA and RNA binding, RNAse activity, and transport into the nucleus, where LF binds specific DNA sequences and activates transcription. The physiological role of LF is still unclear, but it has been suggested to be responsible for primary defence against microbial infections. Here we present evidence that in addition to the above (and below) mentioned functions human milk LF binds ATP with a stoichiometry of 1 mole of nucleotide per mole of the protein and a Kd = 0.3 mM and possesses two sites for interaction with various specific and nonspecific DNA The ATP-binding site is localized in the C-terminal domain of LF (G475-M604), in contrast to the antibacterial and DNA-binding sites, which are located in the N-terminal domain (G1-R31). Binding of ATP by LF leads to dissociation of its oligomeric forms and to a change of the protein’s interaction with polysaccharides, proteins and DNA. The absence of homology of LF with other known transcriptional factors, allosteric changes of the LF interactions with various ligands, and polyfunctional properties of LF distinguish it from other known activators of transcription, and may be very important for untypical functioning of this protein.
Our results taken together with previously observed in literature suggest the existence of a new class of transcriptional activators, those secreted in one cell and taken up by a second target cell in which they are transported to the nucleus and directly activate gene expression.
Introduction
Lactoferrin (LF), a major iron-binding glycoprotein of human milk and other epithelial secretions, is thought to be responsible for primary defence against microbial infections. LF, being relatively iron-unsaturated in human milk, could prevent bacteria from proliferating by removing iron from the surrounding environment. A number of biological functions have been attributed to LF. In particular, a role in the regulation of myelopoiesis has been described by several different groups of investigators. LF was also shown to regulate granulopoiesis, antibody-dependent cytotoxicity, cytokine production, and growth of some cells in vitro. In addition to the above mentioned functions of LF, it is a potent activator of natural killer cells, which may play a role in antitumor defence. Thus, LF is known as an extremely polyfunctional protein, many functions of which appear to be at least in part independent of its iron-binding activity. The nature of such functions remains to be unclear yet. On the over hand, some of these functions may be the result of LF binding to specific DNA sequences. Recent research shows that LF enters the cell from the surrounding environment and is transported into the nucleus where it binds DNA. Specific DNA sequences that can confer lactoferrin-induced gene transcription of a reporter gene have now been identified. In terms of functional activity of LF, the investigation of such DNA- and other polyanions-binding processes induced by lactoferrin would provide a new approach to understanding the mechanisms of multiply regulatory properties of this extremely polyfunctional protein. Clearly, an understanding of these mechanisms is potentially very important but it remains a controversial area of research with inconsistency of results between different investigators and lack of convincing evidence of physiological importance.
Results and discussion
In order to investigate LF, firstly we tried to analyze a possible homology of LF as an activator of transcription with other known transcriptional factors. All sequence analyses were done with the Antheprot for Windows (by G. Deleage) computer programs, using SwissRpot data base. Since, there was no revealed any remarkable homology between LF and other transcriptional factors, it seems reasonable to suppose that LF may be an unique transcriptional regulator. As was shown previously, LF contains the domain having strong homology with a various antimicrobial peptides, for example, magainins from frog skin, cecropins from the hemolymph of insects, and defensins from mammalian. This sequence consists mainly of a loop of 18 amino acid residues formed by a disulfide bond between cystine residues 20 and 37 at the N-terminus of the LF molecule. It seems likely that it is the N-lobe that is responsible for cell receptor recognition by LF and especially for specific sequence DNA binding. But important as this only DNA-binding phenomenon it does not cover the problem of transcriptional regulation mechanism in the case of LF on the whole.
On the basis of these observations, we have proposed that since LF is a small protein (76 kDa), its polyfunctional properties including activation of transcription may result from its existence in several oligomeric forms which have different functions, and that its oligomerization and dissociation are under control of specific ligands such as ATP. Assuming that there may be homology of LF with known ATP-binding enzymes we used computer analysis to clear up this question. A very strong homology of LF with various known ATP- and NADH-binding proteins was revealed. On the next step of the study we have analyzed the interaction of LF with various nonspecific and specific nucleic acids, ATP and a possible relationship between these ligands using different approaches.
 |
 |
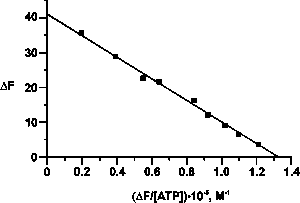
| Figure 1. Determination of the Kd value of the complex between LF and ATP using a Scatchard plot representation of the fluorescence data. | Figure 2. Determination of the Kd value of the complex between LF and nonspecific ON (dT10) using a Scatchard plot representation of the gel retardation data. |
As was shown by different methods, in addition to the above mentioned functions human milk LF binds ATP with a stoichiometry of 1 mole of nucleotide per mole of the protein and a Kd = 0.3 mM (Fig. 1). The ATP-binding site is localized in the C-terminal domain of LF (G475-M604), in contrast to the antibacterial and polyanion-binding sites, which are located in the N-terminal domain (G1-R31). Binding of ATP by LF leads to dissociation of its oligomeric forms and to a change of the protein’s interaction with polysaccharides and proteins.
To estimate the affinity of LF to polyanions (namely DNA, RNA and heparin) several different methods have been used. The Scatchard plots calculated from the gel retardation (Fig. 2) and fluorometric data have shown that LF possesses two different sites for interaction with ss and ds DNA. It is of interest, that both sites of LF can interact with specific and nonspecific ss and ds oligonucleotides (ON). At the same time the affinity of the first site of LF for nonspecific ss and ds dT10 [Kd1(ss)= 1.5×10-6 M and Kd1(ds) = 4.0×10-7 M, respectively] is significantly lower that that in the case of the specific ONs: TAGAAGATCAAA, Kd1(ss)=1.1×10-8 M and Kd1(ds)=8.0×10-8 M. When LF binds the second molecule of nonspecific ON with the affinity about 30 times lower than the first one (Kd2(ss)= 4.5 x10-6 M), its interaction with the first molecule of specific ONs leads to a decrease of the affinity to the second molecule more that three orders of magnitude (Kd2 ~ 1,0 x10-4 M). Thus, there is a very strong antagonistic binding of specific DNA molecules by LF, which is known as phenomena of negative or anti-cooperativity. Moreover, we have revealed using different methods that binding of ATP leads to a change of the LF interaction with both specific and nonspecific DNA and to dramatical conformational changes of the LF molecule.
The further approach provided direct evidence that the LF DNA- and ATP-binding sites do not coincide or overlap and that interaction between these sites has an allosteric nature. The LF became labeled after incubation with the affinity probes for DNA binding sites, the chemically reactive derivatives of specific and nonspecific ONs. The ON- reagents were shown to modify oligopeptide of the N-domain of LF. Thus, DNA-binding site and two different polyanion and antimicrobial domains, which were demonstrated earlier, are located near the N-terminus of LF in a region distinct from its iron-binding site. The question was raised whether the LF site having high affinity for DNA could be the same one that of well known antimicrobial and/or polyanion-binding site. The competitive inhibition experiments have demonstrated that polyanionic RNA, tRNA and heparin inhibit the LF binding with DNA. Therefore, it is reasonable to believe that the DNA-binding site coincides or at least strongly overlaps with well known antimicrobial and polyanion binding sites, located in the N-terminal domain. Taken together, we can conclude that the LF interaction with DNA does not iron-independent and that the site having a high affinity for specific DNA can interact (or at least overlap) with other polyanionic molecules like heparin and RNA or tRNA. At high concentrations of DNA and/or ON the second specific center of LF can also bind polyanionic molecules. Since in the presence of ATP we observed dissociation of the LF tetrameric form into monomers, a decrease of the LF affinity for polysaccharides and proteins, and the influence of ATP on the LF interaction with specific and nonspecific DNA, its reasonable to suggest that such allosteric properties of LF may be very important in the context of its polyfunctional biological functions.
Although the interaction of LF with DNA has been shown to influence on gene transcription, it has yet previously been determined whether any ligands are influenced on physiological function of LF molecule. The finding of such a relationship would provide unequivocal evidence for a biological role of DNA- and ATP-binding properties of LF and firmly establish its status as a functionally active molecule. DNA- and ATP-binding sites may for example play an important role in regulation of the LF binding to cells and for its penetration into cell nuclear.
These results taken together with previously observed in literature suggest the existence of a new class of transcriptional activators, those secreted in one cell and taken up by a second target cell in which they are transported to the nucleus and directly activate gene expression. Moreover, such target gene activation may occur in bacterial cells leads to a previously unknown mechanism of defence against the infecting organisms.
References
- J. He, P. Furmanski, “Sequence specificity and transcriptional activation in the binding of lactoferrin to DNA” Nature 373, 721-724 (1995)
- M. Sousa, J.H. Brock, “Iron in Immunity Cancer and Inflammation” (John Wiley & Sons, Great Britain, 1989)
- D.V. Semenov, T.G. Kanyshkova, A.M. Akimganov, V.N. Buneva, G.A. Nevinsky, “Human milk lactoferrin binds ATP”, Biochemistry (Moscow), accepted for publication
- T.G. Kanyshkova, D.V. Semenov, V.N. Buneva, G.A. Nevinsky, “Localyzation of human milk lactoferrin DNA-binding site”, Bioch. Mol. Biol. Internat., in press.
