LUKASHEV V.A., FROLOV A.S.1, PANIN L.E., GIMAUTDINOVA O.I.
Institute of Biochemistry, Siberian Branch of the Russian Academy of Medical Sciences, Novosibirsk, Russia;
1Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 10 Lavrentiev Ave., Novosibirsk, 630090, Russia;
Keywords: human immunodeficiency virus, glycoprotein, apolipoprotein, protein structure similarity, hydrophobicity profile, packing dencity
T-helper lymphocytes and macrophages are a natural ecological niche for human immunodeficiency virus (HIV) The HIV-infected macrophages don’t die but become a reservoir for the virus, the place where the virus propagates. HIV-infected lymphocytes die causing the development of immunodeficiency. Penetration of the virus into these cells appeared possible due to its reception by the protein CD4, located on the surface of lymphocyte cellular membrane [1].
It is considered that two kinds of the virus were the source of human infection: HIV-1, widespread in chimpanzee, and HIV-2, spread in another monkey species, mangabeys [15]. These viruses for many years remained harmless to humans provoking a mild infection. However, the mutational processes affecting the structure of glycoproteins located on the surface of the viral particle had changed the epitope capable of binding to CD4 [15]. Thus, the virus adapted to an unknown mechanism connected with macrophage and
T-helper function.
Our studies suggest that this mechanism is likely to be the intensification of protein biosynthesis in organs and tissues, including immunocompetent organs and tissues, under the cooperative effect of high-density lipoproteins (HDL) and glucocorticoids. Formation of this mechanism requires necessarily the involvement of macrophages. The mechanism itself includes selective consumption of HDL3 by macrophages and formation of biologically active complex of apolipoprotein (the major protein constituent of HDL) with the reduced forms of steroid hormones, tetrahydro derivatives [7, 8]. On release from the macrophage, this complex enters the nuclei of somatic cells, where it boosts the gene expression [6].
The structural similarity of HIV gp120 and human apolipoprotein A-1, discovered in this study, allowed us to reveal certain mechanisms of HIV antigenic mimicry and manifestation of its pathogenic properties, such as, for instance the weight loss of HIV patients [Fig. 1].
Materials and Methods
The structure similarity of proteins implies first the homology of their primary structures. According to Kimura [2], the rate of functionally permissible amino acid substitutions in proteins that meet one another in the evolutionary pathway depends on the nearest surroundings. This suggestion was confirmed by comparison of the primary and tertiary structures of globular proteins [3, 5]. Basing on this grounds, we developed and applied a method of searching for protein structural similarity to predict their possible functional invariance. To interpret the polyfunctional properties of human apolipoprotein A-1, we used the program POISK. The program implements the scanning of the profile of coordination numbers [9] of the protein under study, similar to hydrophobicity profile [12], over a bank of primary structures of PIR type. In this work, we used PIR, release 34.0 (1996) [13, 14]. The amino acid sequence of human apolipoprotein A-1, isolated from HDP3 [10], was the input data. The coordination numbers reflect the most typical number of the amino acid residues drawn into spatial proximity with a given amino acid residue in the proteins with known 3D structure. A mean value of the coordination number in the neighboring region of N residues around a given residue corresponds to each amino acid residue in the protein primary structure. Thus, any protein can be represented as a sequence of mean coordination numbers of all its amino acid residues. Such curve reflects the exposure extent of amino acids or, vice versa, the extent to which they are shielded in the 3D structure, that is, the protein packing density. Note that the profiles of coordination numbers (packing density) correlate with the protein hydrophobicity profiles, calculated in a different way [9]. The frequency of occurrence of neighboring amino acids with a given amino acid in a protein, expressed as a profile of mean coordination numbers, reflects eventually the structure of the genetic code, which determines the formation of protein functional centers in the course of the evolution. That is why the comparison of proteins according to their profiles of coordination numbers allows the protein regions with similar structure and function to be approximately revealed [5]. To search for such similarity, the profile of coordination numbers of the protein under study is calculated and successively compared with the calculated profiles of peptide segments (30-50 aa) of all the proteins of the bank. The algorithm described is included into the program POISK, which is realized in Fortran-77 and adapted to PC with operational systems MS-DOS, Windows 95, Windows NT, and OS/2-Warp.
Results and Discussion
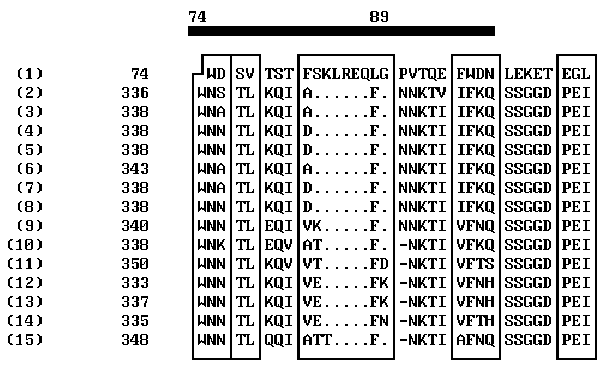
Over ten peptide segments that can be interpreted as sequential antigenic determinants or, more likely, as constituents of spatial antigenic determinants, have been found in HIV env proteins [11]. It is commonly accepted that the major determinants in gp120 interacting with the virus-neutralizing antibodies are located in the region 301-341 aa [11]. Variability of the amino acids flanking this region result in the changes in the antigenic determinants and hinders the development of reliable anti-HIV vaccine. On the other hand, the epitops involved in HIV neutralization are revealed in synthetic peptide fragments of gp120; these epitopes are likely to be disguised on the surface of the natural gp120. One of the reasons of this antigenic disguise can be the structural similarity of these epitops to the fragments of some human proteins. The computer analysis performed allowed us to reveal such structural similarity between gp120 and human apolipoprotein A-1: the segment 74-89 of apolipoprotein A-1 appeared to be similar to antigenically significant region of gp120 (segment 338-354) of several HIV strains (Fig. 2). The homologous regions are indicated on the abscissa. Note that the homology of these regions is very high. This unexpected result aids considerably our understanding of the so-called HIV antigenic mimicry and its ability to escape immune surveillance. It is evident that this peptide site should not be used for construction of artificial vaccines as it fails as an antigen.
The results obtained on the structural similarity of gp120 and human apolipoprotein A-1 in their receptor regions suggest that these proteins enter the cell by a similar mechanism, which is connected with binding to the receptor protein CD4. In this case, HIV may hinder the regular formation in macrophages of a functionally active complex apolipoprotein A-1–tetrahydrocortisol, involved in gene expression boosting, which increases the rate of protein biosynthesis on somatic cells, as was demonstrated in rat hepatocytes [7]. The 71% similarity of the human and rat apolipoprotein A-1 amino acid sequences in the segment 74-89 (90% considering the permissible conservative substitutions [2]) suggests that it is this mechanism that causes the progressive weight loss in HIV patients.
The homology of the peptide segments of human apolipoprotein A-1 and HIV glycoprotein gp120 is not restricted to the data described above. Similar short regions (four-five amino acid residues long) were also found in these proteins as well as the segments of HIV glycoprotein gp41 homologous to human apolipoprotein A-1. The structural similarities of the HIV glycoproteins and human apolipoprotein A-1 that we discovered demonstrate the functional invariance of these proteins and suggest to continue this research direction. If the data obtained are substantiated experimentally, we believe that apolipoprotein A-1 can be a basic object for developing the prototype of a new medication.
References
- A.G. Bukrinskaya and V.M. Zhdanov, “Molecular basis of viral pathogenicity” (Moscow, Meditsina, 1991).
- M. Kimura, “Molecular evolution: the theory of neutrality” (Moscow, Mir, 1985).
- V.A. Lukashev, A.G. Bachinskii, and V.A. Kulichkov, Mol. Biol. (Mosk.), 20, 1192 (1986).
- V.A. Lukashev and V.A. Kulichkov, Biofizika, 35, 236 (1990).
- V.A. Lukashev, “Development of theoretical methods for analysis of functional elements in proteins basing on their invariant structural characteristics” (Cand. Biol. Sci. Dissertation, Novosibirsk, Koltsovo, 1992).
- L.E. Panin, L.M. Polyakov, A.P. Kuz’menko, et al., Biokhimiya, 57, 826 (1992).
- L.E. Panin, I.F. Usynin, O.M. Trubitsyna, et al., Biokhimiya, 59, 249 (1994)
- L.E. Panin, “Functional role of tetrahydro derivatives of steroid hormones”, Proceedings of the Conference “Endocrine mechanisms of the function regulation in norm and pathology” (Novosibirsk, 117, 1997).
- M.A. Rodionov, S.G. Galaktionov, and A.A. Akhrem, Dokl. Akad. Nauk SSSR, 261, 756 (1981).
- H.B. Brewer, T. Fairwell, A. LaRue, R. Ronan, et. al., Biochem. Biophys. Res. Commun., 80, 623 (1978).
- H. Chung, A. Randolph, I. Reardon, et. al., J. Biol. Chem., 257, 2961 (1982).
- J. Kyte and R.F. Doolittle, J. Mol. Biol., 132, 105 (1988).
- A.A. Lukashev, V.M. Blinov, S.S. Bogachev, and N.V. Vlaskin, The 3d All-Union Workshop “Theoretical Investigations and Banks on Molecular Biology and Genetics”, Novosibirsk, July, 18-22, 55 (1988).
- SWISS-PROT Protein Sequence Data Bank. Release 34.0.-October 1996.
- T. Zhu, B.T. Korber, and A.J. Nahmias, Nature, 391, 594 (1998).
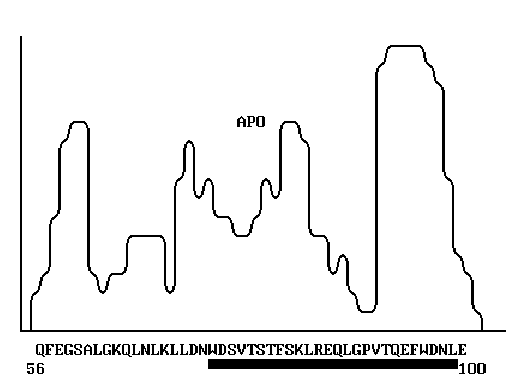

Figure 1. The primary structure of human apolipoprotein A-1 [10]. The segment exhibiting high
homology with the corresponding segment of HIV gp120 is indicated.
(1) APOLIPOPROTEIN HUMAN
ENV_HV1MF (2) 853 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (MFA ISOLATE)
ENV_HV1H3 (3) 856 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (HXB3 ISOLATE)
ENV_HV1PV (4) 856 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (PV22 ISOLATE)
ENV_HV1H2 (5) 856 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (HXB2 ISOLATE)
ENV_HV1BR (6) 861 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF, VIRUS TYPE 1 (BRU ISOLATE)
ENV_HV1B8 (7) 851 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (BH8 ISOLATE)
ENV_HV1B1 (8) 856 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (BH10 ISOLATE)
ENV_HV1A2 (9) 855 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (ARV2/SF2 ISL.)
ENV_HV1S3 (10) 852 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (SF33 ISOLATE)
ENV_HV1RH (11) 865 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (RF/HAT ISOLAT)
ENV_HV1W2 (12) 847 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (WMJ2 ISOLATE)
ENV_HV1W1 (13) 856 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (WMJ1 ISOLATE)
ENV_HV1JR (14) 848 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (JRCSF ISOLATE)
ENV_HV1C4 (15) 868 ENVELOPE POLYPROTEIN GP160 PRECURSOR (CONTAINS: GP120 AND
GP41) (GENE:ENV)-HUMAN IMMUNODEF. VIRUS TYPE 1 (CDC-451 ISOL.)
Figure 2. Comparison of the profiles of coordination numbers of peptide segments 56-100 of human apolipoprotein and 320-365 of HIV gp120.
